Genetic signatures of HIV-1 envelope-mediated bystander apoptosis
- PMID: 24265318
- PMCID: PMC3908386
- DOI: 10.1074/jbc.M113.514018
Genetic signatures of HIV-1 envelope-mediated bystander apoptosis
Abstract
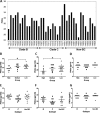
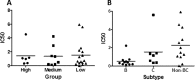
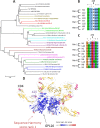
The envelope (Env) glycoprotein of HIV is an important determinant of viral pathogenesis. Several lines of evidence support the role of HIV-1 Env in inducing bystander apoptosis that may be a contributing factor in CD4(+) T cell loss. However, most of the studies testing this phenomenon have been conducted with laboratory-adapted HIV-1 isolates. This raises the question of whether primary Envs derived from HIV-infected patients are capable of inducing bystander apoptosis and whether specific Env signatures are associated with this phenomenon. We developed a high throughput assay to determine the bystander apoptosis inducing activity of a panel of primary Envs. We tested 38 different Envs for bystander apoptosis, virion infectivity, neutralizing antibody sensitivity, and putative N-linked glycosylation sites along with a comprehensive sequence analysis to determine if specific sequence signatures within the viral Env are associated with bystander apoptosis. Our studies show that primary Envs vary considerably in their bystander apoptosis-inducing potential, a phenomenon that correlates inversely with putative N-linked glycosylation sites and positively with virion infectivity. By use of a novel phylogenetic analysis that avoids subtype bias coupled with structural considerations, we found specific residues like Arg-476 and Asn-425 that were associated with differences in bystander apoptosis induction. A specific role of these residues was also confirmed experimentally. These data demonstrate for the first time the potential of primary R5 Envs to mediate bystander apoptosis in CD4(+) T cells. Furthermore, we identify specific genetic signatures within the Env that may be associated with the bystander apoptosis-inducing phenotype.
Keywords: AIDS; Apoptosis; Genetics; HIV; Infectious Diseases; Retrovirus; Virology.
Figures

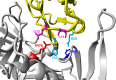
Similar articles
-
HIV ENV glycoprotein-mediated bystander apoptosis depends on expression of the CCR5 co-receptor at the cell surface and ENV fusogenic activity.J Biol Chem. 2011 Oct 21;286(42):36404-13. doi: 10.1074/jbc.M111.281659. Epub 2011 Aug 22. J Biol Chem. 2011. PMID: 21859712 Free PMC article.
-
Host and Viral Factors in HIV-Mediated Bystander Apoptosis.Viruses. 2017 Aug 22;9(8):237. doi: 10.3390/v9080237. Viruses. 2017. PMID: 28829402 Free PMC article. Review.
-
Asn 362 in gp120 contributes to enhanced fusogenicity by CCR5-restricted HIV-1 envelope glycoprotein variants from patients with AIDS.Retrovirology. 2007 Dec 12;4:89. doi: 10.1186/1742-4690-4-89. Retrovirology. 2007. PMID: 18076768 Free PMC article.
-
Identification of Novel Structural Determinants in MW965 Env That Regulate the Neutralization Phenotype and Conformational Masking Potential of Primary HIV-1 Isolates.J Virol. 2018 Feb 12;92(5):e01779-17. doi: 10.1128/JVI.01779-17. Print 2018 Mar 1. J Virol. 2018. PMID: 29237828 Free PMC article.
-
HIV-1 induced bystander apoptosis.Viruses. 2012 Nov 9;4(11):3020-43. doi: 10.3390/v4113020. Viruses. 2012. PMID: 23202514 Free PMC article. Review.
Cited by
-
HIV-1 subtype CRF01_AE and B differ in utilization of low levels of CCR5, Maraviroc susceptibility and potential N-glycosylation sites.Virology. 2017 Dec;512:222-233. doi: 10.1016/j.virol.2017.09.026. Epub 2017 Oct 9. Virology. 2017. PMID: 29020646 Free PMC article.
-
A Partial E3 Deletion in Replication-Defective Adenoviral Vectors Allows for Stable Expression of Potentially Toxic Transgene Products.Hum Gene Ther Methods. 2016 Oct;27(5):187-196. doi: 10.1089/hgtb.2016.044. Epub 2016 Sep 7. Hum Gene Ther Methods. 2016. PMID: 27604324 Free PMC article.
-
HIV-1 envelope glycoproteins isolated from Viremic Non-Progressor individuals are fully functional and cytopathic.Sci Rep. 2019 Apr 3;9(1):5544. doi: 10.1038/s41598-019-42075-3. Sci Rep. 2019. PMID: 30944395 Free PMC article.
-
CCR5 promoter activity correlates with HIV disease progression by regulating CCR5 cell surface expression and CD4 T cell apoptosis.Sci Rep. 2017 Mar 22;7(1):232. doi: 10.1038/s41598-017-00192-x. Sci Rep. 2017. PMID: 28331180 Free PMC article.
-
Combination gene therapy for HIV using a conditional suicidal gene with CCR5 knockout.Virol J. 2021 Jan 30;18(1):31. doi: 10.1186/s12985-021-01501-7. Virol J. 2021. PMID: 33516234 Free PMC article.
References
-
- Douek D. (2007) HIV disease progression. Immune activation, microbes, and a leaky gut. Top. HIV Med. 15, 114–117 - PubMed
-
- Meythaler M., Martinot A., Wang Z., Pryputniewicz S., Kasheta M., Ling B., Marx P. A., O'Neil S., Kaur A. (2009) Differential CD4+ T-lymphocyte apoptosis and bystander T-cell activation in rhesus macaques and sooty mangabeys during acute simian immunodeficiency virus infection. J. Virol. 83, 572–583 - PMC - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Research Materials

