Data, information, knowledge and principle: back to metabolism in KEGG
- PMID: 24214961
- PMCID: PMC3965122
- DOI: 10.1093/nar/gkt1076
Data, information, knowledge and principle: back to metabolism in KEGG
Abstract
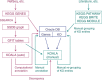
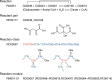
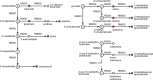
In the hierarchy of data, information and knowledge, computational methods play a major role in the initial processing of data to extract information, but they alone become less effective to compile knowledge from information. The Kyoto Encyclopedia of Genes and Genomes (KEGG) resource (http://www.kegg.jp/ or http://www.genome.jp/kegg/) has been developed as a reference knowledge base to assist this latter process. In particular, the KEGG pathway maps are widely used for biological interpretation of genome sequences and other high-throughput data. The link from genomes to pathways is made through the KEGG Orthology system, a collection of manually defined ortholog groups identified by K numbers. To better automate this interpretation process the KEGG modules defined by Boolean expressions of K numbers have been expanded and improved. Once genes in a genome are annotated with K numbers, the KEGG modules can be computationally evaluated revealing metabolic capacities and other phenotypic features. The reaction modules, which represent chemical units of reactions, have been used to analyze design principles of metabolic networks and also to improve the definition of K numbers and associated annotations. For translational bioinformatics, the KEGG MEDICUS resource has been developed by integrating drug labels (package inserts) used in society.
Figures

Similar articles
-
KEGG for integration and interpretation of large-scale molecular data sets.Nucleic Acids Res. 2012 Jan;40(Database issue):D109-14. doi: 10.1093/nar/gkr988. Epub 2011 Nov 10. Nucleic Acids Res. 2012. PMID: 22080510 Free PMC article.
-
KEGG: new perspectives on genomes, pathways, diseases and drugs.Nucleic Acids Res. 2017 Jan 4;45(D1):D353-D361. doi: 10.1093/nar/gkw1092. Epub 2016 Nov 28. Nucleic Acids Res. 2017. PMID: 27899662 Free PMC article.
-
KEGG for linking genomes to life and the environment.Nucleic Acids Res. 2008 Jan;36(Database issue):D480-4. doi: 10.1093/nar/gkm882. Epub 2007 Dec 12. Nucleic Acids Res. 2008. PMID: 18077471 Free PMC article.
-
Knowledge-Based Analysis of Protein Interaction Networks in Neurodegenerative Diseases.In: Alzate O, editor. Neuroproteomics. Boca Raton (FL): CRC Press/Taylor & Francis; 2010. Chapter 9. In: Alzate O, editor. Neuroproteomics. Boca Raton (FL): CRC Press/Taylor & Francis; 2010. Chapter 9. PMID: 21882442 Free Books & Documents. Review.
-
KEGG as a glycome informatics resource.Glycobiology. 2006 May;16(5):63R-70R. doi: 10.1093/glycob/cwj010. Epub 2005 Jul 13. Glycobiology. 2006. PMID: 16014746 Review.
Cited by
-
Genomic reconstruction of short-chain fatty acid production by the human gut microbiota.Front Mol Biosci. 2022 Aug 11;9:949563. doi: 10.3389/fmolb.2022.949563. eCollection 2022. Front Mol Biosci. 2022. PMID: 36032669 Free PMC article.
-
MRE: a web tool to suggest foreign enzymes for the biosynthesis pathway design with competing endogenous reactions in mind.Nucleic Acids Res. 2016 Jul 8;44(W1):W217-25. doi: 10.1093/nar/gkw342. Epub 2016 Apr 29. Nucleic Acids Res. 2016. PMID: 27131375 Free PMC article.
-
DNA microarray integromics analysis platform.BioData Min. 2015 Jun 25;8:18. doi: 10.1186/s13040-015-0052-6. eCollection 2015. BioData Min. 2015. PMID: 26110022 Free PMC article.
-
Long-term continuous mono-cropping of Macadamia integrifolia greatly affects soil physicochemical properties, rhizospheric bacterial diversity, and metabolite contents.Front Microbiol. 2022 Oct 6;13:952092. doi: 10.3389/fmicb.2022.952092. eCollection 2022. Front Microbiol. 2022. PMID: 36274682 Free PMC article.
-
A key genetic factor for fucosyllactose utilization affects infant gut microbiota development.Nat Commun. 2016 Jun 24;7:11939. doi: 10.1038/ncomms11939. Nat Commun. 2016. PMID: 27340092 Free PMC article.
References
-
- Kanehisa M. Chemical and genomic evolution of enzyme-catalyzed reaction networks. FEBS Lett. 2013;587:2731–2737. - PubMed

