Epstein-Barr virus nuclear antigen leader protein localizes to promoters and enhancers with cell transcription factors and EBNA2
- PMID: 24167291
- PMCID: PMC3832032
- DOI: 10.1073/pnas.1317608110
Epstein-Barr virus nuclear antigen leader protein localizes to promoters and enhancers with cell transcription factors and EBNA2
Abstract
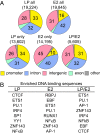
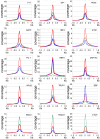
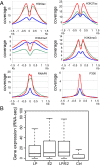
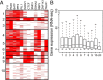
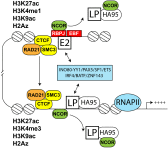
Epstein-Barr virus (EBV) nuclear antigens EBNALP (LP) and EBNA2 (E2) are coexpressed in EBV-infected B lymphocytes and are critical for lymphoblastoid cell line outgrowth. LP removes NCOR and RBPJ repressive complexes from promoters, enhancers, and matrix-associated deacetylase bodies, whereas E2 activates transcription from distal enhancers. LP ChIP-seq analyses identified 19,224 LP sites of which ~50% were ± 2 kb of a transcriptional start site. LP sites were enriched for B-cell transcription factors (TFs), YY1, SP1, PAX5, BATF, IRF4, ETS1, RAD21, PU.1, CTCF, RBPJ, ZNF143, SMC3, NFκB, TBLR, and EBF. E2 sites were also highly enriched for LP-associated cell TFs and were more highly occupied by RBPJ and EBF. LP sites were highly marked by H3K4me3, H3K27ac, H2Az, H3K9ac, RNAPII, and P300, indicative of activated transcription. LP sites were 29% colocalized with E2 (LP/E2). LP/E2 sites were more similar to LP than to E2 sites in associated cell TFs, RNAPII, P300, and histone H3K4me3, H3K9ac, H3K27ac, and H2Az occupancy, and were more highly transcribed than LP or E2 sites. Gene affected by CTCF and LP cooccupancy were more highly expressed than genes affected by CTCF alone. LP was at myc enhancers and promoters and of MYC regulated ccnd2, 23 med complex components, and MYC regulated cell survival genes, igf2r and bcl2. These data implicate LP and associated TFs and DNA looping factors CTCF, RAD21, SMC3, and YY1/INO80 chromatin-remodeling complexes in repressor depletion and gene activation necessary for lymphoblastoid cell line growth and survival.
Keywords: gene expression; genome-wide ChIP-seq analysis.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
Similar articles
-
Epstein-Barr Virus Nuclear Antigen Leader Protein Coactivates EP300.J Virol. 2018 Apr 13;92(9):e02155-17. doi: 10.1128/JVI.02155-17. Print 2018 May 1. J Virol. 2018. PMID: 29467311 Free PMC article.
-
EBV nuclear antigen EBNALP dismisses transcription repressors NCoR and RBPJ from enhancers and EBNA2 increases NCoR-deficient RBPJ DNA binding.Proc Natl Acad Sci U S A. 2011 May 10;108(19):7808-13. doi: 10.1073/pnas.1104991108. Epub 2011 Apr 25. Proc Natl Acad Sci U S A. 2011. PMID: 21518914 Free PMC article.
-
Epstein-Barr virus exploits intrinsic B-lymphocyte transcription programs to achieve immortal cell growth.Proc Natl Acad Sci U S A. 2011 Sep 6;108(36):14902-7. doi: 10.1073/pnas.1108892108. Epub 2011 Jul 11. Proc Natl Acad Sci U S A. 2011. PMID: 21746931 Free PMC article.
-
Both Epstein-Barr viral nuclear antigen 2 (EBNA2) and activated Notch1 transactivate genes by interacting with the cellular protein RBP-J kappa.Immunobiology. 1997 Dec;198(1-3):299-306. doi: 10.1016/s0171-2985(97)80050-2. Immunobiology. 1997. PMID: 9442401 Review.
-
EBNA2 and Its Coactivator EBNA-LP.Curr Top Microbiol Immunol. 2015;391:35-59. doi: 10.1007/978-3-319-22834-1_2. Curr Top Microbiol Immunol. 2015. PMID: 26428371 Review.
Cited by
-
Phase Separation of Epstein-Barr Virus EBNA2 and Its Coactivator EBNALP Controls Gene Expression.J Virol. 2020 Mar 17;94(7):e01771-19. doi: 10.1128/JVI.01771-19. Print 2020 Mar 17. J Virol. 2020. PMID: 31941785 Free PMC article.
-
Epstein-Barr Virus B Cell Growth Transformation: The Nuclear Events.Viruses. 2023 Mar 24;15(4):832. doi: 10.3390/v15040832. Viruses. 2023. PMID: 37112815 Free PMC article. Review.
-
Epstein-Barr virus latency: current and future perspectives.Curr Opin Virol. 2015 Oct;14:138-44. doi: 10.1016/j.coviro.2015.09.007. Curr Opin Virol. 2015. PMID: 26453799 Free PMC article. Review.
-
The Epstein-Barr Virus Regulome in Lymphoblastoid Cells.Cell Host Microbe. 2017 Oct 11;22(4):561-573.e4. doi: 10.1016/j.chom.2017.09.001. Cell Host Microbe. 2017. PMID: 29024646 Free PMC article.
-
A new framework for host-pathogen interaction research.Front Immunol. 2022 Dec 15;13:1066733. doi: 10.3389/fimmu.2022.1066733. eCollection 2022. Front Immunol. 2022. PMID: 36591248 Free PMC article.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

