Crystal structure of the gamma-2 herpesvirus LANA DNA binding domain identifies charged surface residues which impact viral latency
- PMID: 24146618
- PMCID: PMC3798461
- DOI: 10.1371/journal.ppat.1003673
Crystal structure of the gamma-2 herpesvirus LANA DNA binding domain identifies charged surface residues which impact viral latency
Abstract
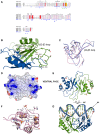
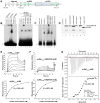
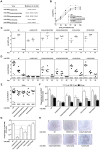
Latency-associated nuclear antigen (LANA) mediates γ2-herpesvirus genome persistence and regulates transcription. We describe the crystal structure of the murine gammaherpesvirus-68 LANA C-terminal domain at 2.2 Å resolution. The structure reveals an alpha-beta fold that assembles as a dimer, reminiscent of Epstein-Barr virus EBNA1. A predicted DNA binding surface is present and opposite this interface is a positive electrostatic patch. Targeted DNA recognition substitutions eliminated DNA binding, while certain charged patch mutations reduced bromodomain protein, BRD4, binding. Virus containing LANA abolished for DNA binding was incapable of viable latent infection in mice. Virus with mutations at the charged patch periphery exhibited substantial deficiency in expansion of latent infection, while central region substitutions had little effect. This deficiency was independent of BRD4. These results elucidate the LANA DNA binding domain structure and reveal a unique charged region that exerts a critical role in viral latent infection, likely acting through a host cell protein(s).
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures


Similar articles
-
The Kaposi Sarcoma Herpesvirus Latency-associated Nuclear Antigen DNA Binding Domain Dorsal Positive Electrostatic Patch Facilitates DNA Replication and Episome Persistence.J Biol Chem. 2015 Nov 20;290(47):28084-28096. doi: 10.1074/jbc.M115.674622. Epub 2015 Sep 29. J Biol Chem. 2015. PMID: 26420481 Free PMC article.
-
A structural basis for BRD2/4-mediated host chromatin interaction and oligomer assembly of Kaposi sarcoma-associated herpesvirus and murine gammaherpesvirus LANA proteins.PLoS Pathog. 2013;9(10):e1003640. doi: 10.1371/journal.ppat.1003640. Epub 2013 Oct 17. PLoS Pathog. 2013. PMID: 24146614 Free PMC article.
-
Mutational analysis of the latency-associated nuclear antigen DNA-binding domain of Kaposi's sarcoma-associated herpesvirus reveals structural conservation among gammaherpesvirus origin-binding proteins.J Gen Virol. 2010 Sep;91(Pt 9):2203-15. doi: 10.1099/vir.0.020958-0. Epub 2010 May 19. J Gen Virol. 2010. PMID: 20484563 Free PMC article.
-
Kaposi's Sarcoma-Associated Herpesvirus Latency-Associated Nuclear Antigen: Replicating and Shielding Viral DNA during Viral Persistence.J Virol. 2017 Jun 26;91(14):e01083-16. doi: 10.1128/JVI.01083-16. Print 2017 Jul 15. J Virol. 2017. PMID: 28446671 Free PMC article. Review.
-
Chromatin organization of gammaherpesvirus latent genomes.Biochim Biophys Acta. 2010 Mar-Apr;1799(3-4):236-45. doi: 10.1016/j.bbagrm.2009.10.004. Epub 2009 Oct 22. Biochim Biophys Acta. 2010. PMID: 19853673 Free PMC article. Review.
Cited by
-
Recent advances in understanding Kaposi's sarcoma-associated herpesvirus.F1000Res. 2016 Apr 25;5:F1000 Faculty Rev-740. doi: 10.12688/f1000research.7612.1. eCollection 2016. F1000Res. 2016. PMID: 27158465 Free PMC article. Review.
-
Conquering the Host: Determinants of Pathogenesis Learned from Murine Gammaherpesvirus 68.Annu Rev Virol. 2021 Sep 29;8(1):349-371. doi: 10.1146/annurev-virology-011921-082615. Annu Rev Virol. 2021. PMID: 34586873 Free PMC article.
-
The Kaposi Sarcoma Herpesvirus Latency-associated Nuclear Antigen DNA Binding Domain Dorsal Positive Electrostatic Patch Facilitates DNA Replication and Episome Persistence.J Biol Chem. 2015 Nov 20;290(47):28084-28096. doi: 10.1074/jbc.M115.674622. Epub 2015 Sep 29. J Biol Chem. 2015. PMID: 26420481 Free PMC article.
-
In Vivo Persistence of Chimeric Virus after Substitution of the Kaposi's Sarcoma-Associated Herpesvirus LANA DNA Binding Domain with That of Murid Herpesvirus 4.J Virol. 2018 Oct 12;92(21):e01251-18. doi: 10.1128/JVI.01251-18. Print 2018 Nov 1. J Virol. 2018. PMID: 30111565 Free PMC article.
-
Structural Basis for Cooperative Binding of EBNA1 to the Epstein-Barr Virus Dyad Symmetry Minimal Origin of Replication.J Virol. 2019 Sep 30;93(20):e00487-19. doi: 10.1128/JVI.00487-19. Print 2019 Oct 15. J Virol. 2019. PMID: 31142669 Free PMC article.
References
-
- Simas JP, Efstathiou S (1998) Murine gammaherpesvirus 68: a model for the study of gammaherpesvirus pathogenesis. Trends Microbiol 6: 276–282. - PubMed
-
- Sunil-Chandra NP, Efstathiou S, Nash AA (1992) Murine gammaherpesvirus 68 establishes a latent infection in mouse B lymphocytes in vivo. J Gen Virol 73 Pt 12: 3275–3279. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

