Microbiota-mediated colonization resistance against intestinal pathogens
- PMID: 24096337
- PMCID: PMC4194195
- DOI: 10.1038/nri3535
Microbiota-mediated colonization resistance against intestinal pathogens
Abstract
Commensal bacteria inhabit mucosal and epidermal surfaces in mice and humans, and have effects on metabolic and immune pathways in their hosts. Recent studies indicate that the commensal microbiota can be manipulated to prevent and even to cure infections that are caused by pathogenic bacteria, particularly pathogens that are broadly resistant to antibiotics, such as vancomycin-resistant Enterococcus faecium, Gram-negative Enterobacteriaceae and Clostridium difficile. In this Review, we discuss how immune- mediated colonization resistance against antibiotic-resistant intestinal pathogens is influenced by the composition of the commensal microbiota. We also review recent advances characterizing the ability of different commensal bacterial families, genera and species to restore colonization resistance to intestinal pathogens in antibiotic-treated hosts.
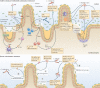
Figures

Comment in
-
Biochemical study of monkeypox zoonotic disease associated human skin dysbiosis and soft tissue injury - Correspondence.Int J Surg. 2022 Nov;107:106977. doi: 10.1016/j.ijsu.2022.106977. Epub 2022 Nov 1. Int J Surg. 2022. PMID: 36332786 Free PMC article. No abstract available.
Similar articles
-
Cooperating Commensals Restore Colonization Resistance to Vancomycin-Resistant Enterococcus faecium.Cell Host Microbe. 2017 May 10;21(5):592-602.e4. doi: 10.1016/j.chom.2017.04.002. Cell Host Microbe. 2017. PMID: 28494240 Free PMC article.
-
The Roles of Inflammation, Nutrient Availability and the Commensal Microbiota in Enteric Pathogen Infection.Microbiol Spectr. 2015 Jun;3(3). doi: 10.1128/microbiolspec.MBP-0008-2014. Microbiol Spectr. 2015. PMID: 26185088
-
Resurrecting the intestinal microbiota to combat antibiotic-resistant pathogens.Science. 2016 Apr 29;352(6285):535-8. doi: 10.1126/science.aad9382. Science. 2016. PMID: 27126035 Free PMC article. Review.
-
Antibiotic-Driven Dysbiosis Mediates Intraluminal Agglutination and Alternative Segregation of Enterococcus faecium from the Intestinal Epithelium.mBio. 2015 Nov 10;6(6):e01346-15. doi: 10.1128/mBio.01346-15. mBio. 2015. PMID: 26556272 Free PMC article.
-
Immune-Microbiota Interplay and Colonization Resistance in Infection.Mol Cell. 2020 May 21;78(4):597-613. doi: 10.1016/j.molcel.2020.03.001. Epub 2020 Mar 23. Mol Cell. 2020. PMID: 32208169 Review.
Cited by
-
Gut microbial community in proboscis monkeys (Nasalis larvatus): implications for effects of geographical and social factors.R Soc Open Sci. 2024 Jul 24;11(7):231756. doi: 10.1098/rsos.231756. eCollection 2024 Jul. R Soc Open Sci. 2024. PMID: 39050721 Free PMC article.
-
Early life microbial colonization of the gut and intestinal development differ between genetically divergent broiler lines.BMC Genomics. 2015 May 28;16(1):418. doi: 10.1186/s12864-015-1646-6. BMC Genomics. 2015. PMID: 26017153 Free PMC article.
-
Unveiling the mechanism of essential oil action against skin pathogens: from ancient wisdom to modern science.Arch Microbiol. 2024 Jul 10;206(8):347. doi: 10.1007/s00203-024-03986-6. Arch Microbiol. 2024. PMID: 38985339 Review.
-
An ecological framework to understand the efficacy of fecal microbiota transplantation.Nat Commun. 2020 Jul 3;11(1):3329. doi: 10.1038/s41467-020-17180-x. Nat Commun. 2020. PMID: 32620839 Free PMC article.
-
Correlation between the gut microbiota characteristics of hosts with severe acute pancreatitis and secondary intra-abdominal infection.Front Med (Lausanne). 2024 Aug 21;11:1409409. doi: 10.3389/fmed.2024.1409409. eCollection 2024. Front Med (Lausanne). 2024. PMID: 39234039 Free PMC article.
References
-
- Sensakovic JW, Smith LG. Oral antibiotic treatment of infectious diseases. Med. Clin. North. Am. 2001;85:115–123. vii. - PubMed
-
- Diehl GE, et al. Microbiota restricts trafficking of bacteria to mesenteric lymph nodes by CX3CR1hi cells. Nature. 2013;494:116–120. [This study shows that the normal microbiota stimulates MYD88-dependent pathways to restrict the delivery of commensal bacteria to the mesenteric lymph nodes.] - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

