Evolution of the JAK-STAT pathway
- PMID: 24058787
- PMCID: PMC3670263
- DOI: 10.4161/jkst.22756
Evolution of the JAK-STAT pathway
Abstract
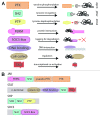
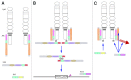
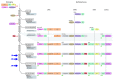
The JAK-STAT pathway represents a finely tuned orchestra capable of rapidly facilitating an exquisite symphony of responses from a complex array of extracellular signals. This review explores the evolution of the JAK-STAT pathway: the origins of the individual domains from which it is constructed, the formation of individual components from these basic building blocks, the assembly of the components into a functional pathway, and the subsequent reiteration of this basic template to fulfill a variety of roles downstream of cytokine receptors.
Keywords: JAK; SHP; SOCS; STAT; evolution.
Figures
Similar articles
-
Signaling via the CytoR/JAK/STAT/SOCS pathway: Emergence during evolution.Mol Immunol. 2016 Mar;71:166-175. doi: 10.1016/j.molimm.2016.02.002. Epub 2016 Feb 18. Mol Immunol. 2016. PMID: 26897340
-
Insulin-like growth factor-I receptor signal transduction and the Janus Kinase/Signal Transducer and Activator of Transcription (JAK-STAT) pathway.Biofactors. 2009 Jan-Feb;35(1):76-81. doi: 10.1002/biof.20. Biofactors. 2009. PMID: 19319849 Review.
-
Suppressors of cytokine signaling abrogate diabetic nephropathy.J Am Soc Nephrol. 2010 May;21(5):763-72. doi: 10.1681/ASN.2009060625. Epub 2010 Feb 25. J Am Soc Nephrol. 2010. PMID: 20185635 Free PMC article.
-
[Negative regulation of the JAK/STAT: pathway implication in tumorigenesis].Bull Cancer. 2005 Oct;92(10):845-57. Bull Cancer. 2005. PMID: 16266868 French.
-
Jak/STAT/SOCS signaling circuits and associated cytokine-mediated inflammation and hypertrophy in the heart.Shock. 2006 Sep;26(3):226-34. doi: 10.1097/01.shk.0000226341.32786.b9. Shock. 2006. PMID: 16912647 Review.
Cited by
-
JAK2 as Predictor of Therapeutic Response in Patients with Chronic Myeloid Leukemia Treated with Imatinib.Dis Markers. 2024 Apr 12;2024:2906566. doi: 10.1155/2024/2906566. eCollection 2024. Dis Markers. 2024. PMID: 38716474 Free PMC article.
-
The Role of the JAK-STAT Pathway in Childhood B-Cell Acute Lymphoblastic Leukemia.Int J Mol Sci. 2024 Jun 21;25(13):6844. doi: 10.3390/ijms25136844. Int J Mol Sci. 2024. PMID: 38999955 Free PMC article. Review.
-
Transcriptional, Epigenetic and Pharmacological Control of JAK/STAT Pathway in NK Cells.Front Immunol. 2019 Oct 17;10:2456. doi: 10.3389/fimmu.2019.02456. eCollection 2019. Front Immunol. 2019. PMID: 31681330 Free PMC article. Review.
-
JAK-STAT signaling regulation of chicken embryonic stem cell differentiation into male germ cells.In Vitro Cell Dev Biol Anim. 2017 Sep;53(8):728-743. doi: 10.1007/s11626-017-0167-9. Epub 2017 Jun 8. In Vitro Cell Dev Biol Anim. 2017. PMID: 28597334
-
Annona squamosa Fruit Extract Ameliorates Lead Acetate-Induced Testicular Injury by Modulating JAK-1/STAT-3/SOCS-1 Signaling in Male Rats.Int J Mol Sci. 2024 May 20;25(10):5562. doi: 10.3390/ijms25105562. Int J Mol Sci. 2024. PMID: 38791600 Free PMC article.
References
Publication types
LinkOut - more resources
Full Text Sources
Other Literature Sources
