An alteration in ELMOD3, an Arl2 GTPase-activating protein, is associated with hearing impairment in humans
- PMID: 24039609
- PMCID: PMC3764207
- DOI: 10.1371/journal.pgen.1003774
An alteration in ELMOD3, an Arl2 GTPase-activating protein, is associated with hearing impairment in humans
Abstract
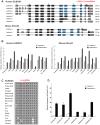
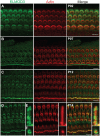
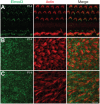
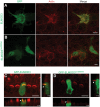
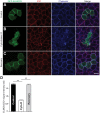
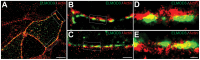
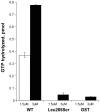
Exome sequencing coupled with homozygosity mapping was used to identify a transition mutation (c.794T>C; p.Leu265Ser) in ELMOD3 at the DFNB88 locus that is associated with nonsyndromic deafness in a large Pakistani family, PKDF468. The affected individuals of this family exhibited pre-lingual, severe-to-profound degrees of mixed hearing loss. ELMOD3 belongs to the engulfment and cell motility (ELMO) family, which consists of six paralogs in mammals. Several members of the ELMO family have been shown to regulate a subset of GTPases within the Ras superfamily. However, ELMOD3 is a largely uncharacterized protein that has no previously known biochemical activities. We found that in rodents, within the sensory epithelia of the inner ear, ELMOD3 appears most pronounced in the stereocilia of cochlear hair cells. Fluorescently tagged ELMOD3 co-localized with the actin cytoskeleton in MDCK cells and actin-based microvilli of LLC-PK1-CL4 epithelial cells. The p.Leu265Ser mutation in the ELMO domain impaired each of these activities. Super-resolution imaging revealed instances of close association of ELMOD3 with actin at the plasma membrane of MDCK cells. Furthermore, recombinant human GST-ELMOD3 exhibited GTPase activating protein (GAP) activity against the Arl2 GTPase, which was completely abolished by the p.Leu265Ser mutation. Collectively, our data provide the first insights into the expression and biochemical properties of ELMOD3 and highlight its functional links to sound perception and actin cytoskeleton.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures

Similar articles
-
Elmod3 knockout leads to progressive hearing loss and abnormalities in cochlear hair cell stereocilia.Hum Mol Genet. 2019 Dec 15;28(24):4103-4112. doi: 10.1093/hmg/ddz240. Hum Mol Genet. 2019. PMID: 31628468 Free PMC article.
-
Mutations of the mouse ELMO domain containing 1 gene (Elmod1) link small GTPase signaling to actin cytoskeleton dynamics in hair cell stereocilia.PLoS One. 2012;7(4):e36074. doi: 10.1371/journal.pone.0036074. Epub 2012 Apr 27. PLoS One. 2012. PMID: 22558334 Free PMC article.
-
ELMOD3, a novel causative gene, associated with human autosomal dominant nonsyndromic and progressive hearing loss.Hum Genet. 2018 Apr;137(4):329-342. doi: 10.1007/s00439-018-1885-0. Epub 2018 Apr 30. Hum Genet. 2018. PMID: 29713870
-
Hair cells, plasma membrane Ca²⁺ ATPase and deafness.Int J Biochem Cell Biol. 2012 May;44(5):679-83. doi: 10.1016/j.biocel.2012.02.006. Epub 2012 Feb 13. Int J Biochem Cell Biol. 2012. PMID: 22349217 Review.
-
Stereocilia morphogenesis and maintenance through regulation of actin stability.Semin Cell Dev Biol. 2017 May;65:88-95. doi: 10.1016/j.semcdb.2016.08.017. Epub 2016 Aug 23. Semin Cell Dev Biol. 2017. PMID: 27565685 Free PMC article. Review.
Cited by
-
ARNSHL gene identification: past, present and future.Mol Genet Genomics. 2022 Sep;297(5):1185-1193. doi: 10.1007/s00438-022-01926-x. Epub 2022 Jul 23. Mol Genet Genomics. 2022. PMID: 35869994 Review.
-
PDZD7 and hearing loss: More than just a modifier.Am J Med Genet A. 2015 Dec;167A(12):2957-65. doi: 10.1002/ajmg.a.37274. Epub 2015 Sep 29. Am J Med Genet A. 2015. PMID: 26416264 Free PMC article.
-
Mutations of human NARS2, encoding the mitochondrial asparaginyl-tRNA synthetase, cause nonsyndromic deafness and Leigh syndrome.PLoS Genet. 2015 Mar 25;11(3):e1005097. doi: 10.1371/journal.pgen.1005097. eCollection 2015 Mar. PLoS Genet. 2015. PMID: 25807530 Free PMC article.
-
Effect of the N-Terminal Helix and Nucleotide Loading on the Membrane and Effector Binding of Arl2/3.Biophys J. 2015 Oct 20;109(8):1619-29. doi: 10.1016/j.bpj.2015.08.033. Biophys J. 2015. PMID: 26488653 Free PMC article.
-
Autosomal recessive non-syndromic hearing loss genes in Pakistan during the previous three decades.J Cell Mol Med. 2024 Apr;28(8):e18119. doi: 10.1111/jcmm.18119. J Cell Mol Med. 2024. PMID: 38534090 Free PMC article. Review.
References
-
- Brown SD, Hardisty-Hughes RE, Mburu P (2008) Quiet as a mouse: dissecting the molecular and genetic basis of hearing. Nature reviews Genetics 9: 277–290. - PubMed
-
- Frolenkov GI, Belyantseva IA, Friedman TB, Griffith AJ (2004) Genetic insights into the morphogenesis of inner ear hair cells. Nature reviews Genetics 5: 489–498. - PubMed
-
- Richardson GP, de Monvel JB, Petit C (2011) How the genetics of deafness illuminates auditory physiology. Annual review of physiology 73: 311–334. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

