Comparing CDRH3 diversity captured from secondary lymphoid organs for the generation of recombinant human antibodies
- PMID: 23924800
- PMCID: PMC3851222
- DOI: 10.4161/mabs.25592
Comparing CDRH3 diversity captured from secondary lymphoid organs for the generation of recombinant human antibodies
Abstract
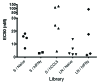
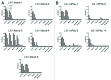
The plasticity of natural immunoglobulin repertoires can be exploited for the generation of phage display libraries. Secondary lymphoid organs, such as the spleen and the lymph nodes, constitute interesting sources of diversity because they are rich in B cells, part of which can be affinity matured. These organs, however, differ in their anatomical structure, reflecting the different fluids they drain, which affects the B cell repertoires. The CDRH3 repertoires from these organs, extracted from naïve or immunized mice, were compared in the context of phage display libraries using human antibody framework families. Deep sequencing analysis revealed that all libraries displayed different CDRH3 repertoires, but the one derived from lymph nodes of naïve mice was the most diverse. Library performance was assessed by in vitro selection. For both organs, immunization increased substantially the frequency of molecules able to bind to the immunogen. The library derived from lymph nodes from naïve mice, however, was the most effective in generating diverse and high affinity candidates. These results illustrate that the use of a biased CDRH3 repertoire increases the performance of libraries, but reduces the clonal diversity, which may be detrimental for certain strategies.
Keywords: antibody repertoire; immune library; next generation sequencing; phage display; scFv; secondary lymphoid organs.
Figures
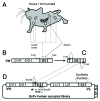
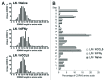
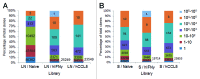
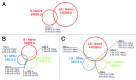
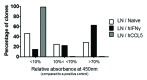
Similar articles
-
Transferring the characteristics of naturally occurring and biased antibody repertoires to human antibody libraries by trapping CDRH3 sequences.PLoS One. 2012;7(8):e43471. doi: 10.1371/journal.pone.0043471. Epub 2012 Aug 24. PLoS One. 2012. PMID: 22937053 Free PMC article.
-
A novel heavy domain antibody library with functionally optimized complementarity determining regions.PLoS One. 2013 Oct 8;8(10):e76834. doi: 10.1371/journal.pone.0076834. eCollection 2013. PLoS One. 2013. PMID: 24116173 Free PMC article.
-
Somatic diversity of the immunoglobulin repertoire is controlled in an isotype-specific manner.Eur J Immunol. 2001 Aug;31(8):2319-30. doi: 10.1002/1521-4141(200108)31:8<2319::aid-immu2319>3.0.co;2-t. Eur J Immunol. 2001. PMID: 11477544
-
Diversity in the Cow Ultralong CDR H3 Antibody Repertoire.Front Immunol. 2018 Jun 4;9:1262. doi: 10.3389/fimmu.2018.01262. eCollection 2018. Front Immunol. 2018. PMID: 29915599 Free PMC article. Review.
-
Synthetic antibodies: concepts, potential and practical considerations.Methods. 2012 Aug;57(4):486-98. doi: 10.1016/j.ymeth.2012.06.012. Epub 2012 Jun 27. Methods. 2012. PMID: 22750306 Review.
Cited by
-
VDJtools: Unifying Post-analysis of T Cell Receptor Repertoires.PLoS Comput Biol. 2015 Nov 25;11(11):e1004503. doi: 10.1371/journal.pcbi.1004503. eCollection 2015 Nov. PLoS Comput Biol. 2015. PMID: 26606115 Free PMC article.
-
Discovery of high affinity anti-ricin antibodies by B cell receptor sequencing and by yeast display of combinatorial VH:VL libraries from immunized animals.MAbs. 2016 Aug-Sep;8(6):1035-44. doi: 10.1080/19420862.2016.1190059. Epub 2016 May 25. MAbs. 2016. PMID: 27224530 Free PMC article.
-
High-throughput sequencing enhanced phage display enables the identification of patient-specific epitope motifs in serum.Sci Rep. 2015 Aug 6;5:12913. doi: 10.1038/srep12913. Sci Rep. 2015. PMID: 26246327 Free PMC article.
-
SAROTUP: a suite of tools for finding potential target-unrelated peptides from phage display data.Int J Biol Sci. 2019 Jun 2;15(7):1452-1459. doi: 10.7150/ijbs.31957. eCollection 2019. Int J Biol Sci. 2019. PMID: 31337975 Free PMC article.
-
Phage display peptide libraries: deviations from randomness and correctives.Nucleic Acids Res. 2018 May 18;46(9):e52. doi: 10.1093/nar/gky077. Nucleic Acids Res. 2018. PMID: 29420788 Free PMC article.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
