Structural basis for R-spondin recognition by LGR4/5/6 receptors
- PMID: 23756652
- PMCID: PMC3701189
- DOI: 10.1101/gad.219360.113
Structural basis for R-spondin recognition by LGR4/5/6 receptors
Abstract
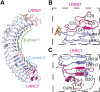
The R-spondin (RSPO) family of secreted proteins (RSPO1-RSPO4) has pleiotropic functions in development and stem cell growth by strongly enhancing Wnt pathway activation. Recently, leucine-rich repeat-containing G-protein-coupled receptor 4 (LGR4), LGR5, and LGR6 have been identified as receptors for RSPOs. Here we report the complex structure of the LGR4 extracellular domain (ECD) with the RSPO1 N-terminal fragment (RSPO1-2F) containing two adjacent furin-like cysteine-rich domains (FU-CRDs). The LGR4-ECD adopts the anticipated TLR horseshoe structure and uses its concave surface close to the N termini to bind RSPO1-2F. Both the FU-CRD1 and FU-CRD2 domains of RSPO1 contribute to LGR4 interaction, and binding and cellular assays identified critical RSPO1 residues for its biological activities. Our results define the molecular mechanism by which the LGR4/5/6 receptors recognize RSPOs and also provide structural insights into the signaling difference between the LGR4/5/6 receptors and other members in the LGR family.
Keywords: Wnt signal; complex; ligand/receptor interaction.
Figures
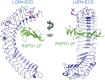


Similar articles
-
Crystal structure of LGR4-Rspo1 complex: insights into the divergent mechanisms of ligand recognition by leucine-rich repeat G-protein-coupled receptors (LGRs).J Biol Chem. 2015 Jan 23;290(4):2455-65. doi: 10.1074/jbc.M114.599134. Epub 2014 Dec 5. J Biol Chem. 2015. PMID: 25480784 Free PMC article.
-
Reconstitution of R-spondin:LGR4:ZNRF3 adult stem cell growth factor signaling complexes with recombinant proteins produced in Escherichia coli.Biochemistry. 2013 Oct 15;52(41):7295-304. doi: 10.1021/bi401090h. Epub 2013 Oct 3. Biochemistry. 2013. PMID: 24050775 Free PMC article.
-
Structure of stem cell growth factor R-spondin 1 in complex with the ectodomain of its receptor LGR5.Cell Rep. 2013 Jun 27;3(6):1885-92. doi: 10.1016/j.celrep.2013.06.009. Cell Rep. 2013. PMID: 23809763
-
R-spondins: Multi-mode WNT signaling regulators in adult stem cells.Int J Biochem Cell Biol. 2019 Jan;106:26-34. doi: 10.1016/j.biocel.2018.11.005. Epub 2018 Nov 12. Int J Biochem Cell Biol. 2019. PMID: 30439551 Review.
-
Structure and function of LGR5: an enigmatic G-protein coupled receptor marking stem cells.Protein Sci. 2014 May;23(5):551-65. doi: 10.1002/pro.2446. Epub 2014 Mar 19. Protein Sci. 2014. PMID: 24677446 Free PMC article. Review.
Cited by
-
A novel function of R-spondin1 in regulating estrogen receptor expression independent of Wnt/β-catenin signaling.Elife. 2020 Aug 4;9:e56434. doi: 10.7554/eLife.56434. Elife. 2020. PMID: 32749219 Free PMC article.
-
LGR4: A New Receptor Member in Endocrine and Metabolic Diseases.Endocr Rev. 2023 Jul 11;44(4):647-667. doi: 10.1210/endrev/bnad003. Endocr Rev. 2023. PMID: 36791020 Free PMC article. Review.
-
Protease associated domain of RNF43 is not necessary for the suppression of Wnt/β-catenin signaling in human cells.Cell Commun Signal. 2020 Jun 11;18(1):91. doi: 10.1186/s12964-020-00559-0. Cell Commun Signal. 2020. PMID: 32527265 Free PMC article.
-
Canonical WNT/β-Catenin Signaling Activated by WNT9b and RSPO2 Cooperation Regulates Facial Morphogenesis in Mice.Front Cell Dev Biol. 2020 May 8;8:264. doi: 10.3389/fcell.2020.00264. eCollection 2020. Front Cell Dev Biol. 2020. PMID: 32457899 Free PMC article.
-
Crystal structure of LGR4-Rspo1 complex: insights into the divergent mechanisms of ligand recognition by leucine-rich repeat G-protein-coupled receptors (LGRs).J Biol Chem. 2015 Jan 23;290(4):2455-65. doi: 10.1074/jbc.M114.599134. Epub 2014 Dec 5. J Biol Chem. 2015. PMID: 25480784 Free PMC article.
References
-
- Adams PD, Grosse-Kunstleve RW, Hung LW, Ioerger TR, McCoy AJ, Moriarty NW, Read RJ, Sacchettini JC, Sauter NK, Terwilliger TC 2002. PHENIX: Building new software for automated crystallographic structure determination. Acta Crystallogr D Biol Crystallogr 58: 1948–1954 - PubMed
-
- Barker N, Clevers H 2010. Leucine-rich repeat-containing G-protein-coupled receptors as markers of adult stem cells. Gastroenterology 138: 1681–1696 - PubMed
-
- Birchmeier W 2011. Stem cells: Orphan receptors find a home. Nature 476: 287–288 - PubMed
-
- Blaydon D, Ishii Y, O'Toole E, Unsworth H, Teh M, Ruschendorf F, Sinclair C, Hopsu-Havu V, Tidman N, Moss C, et al. 2006. The gene encoding R-spondin 4 (RSPO4), a secreted protein implicated in Wnt signaling, is mutated in inherited anonychia. Nat Genet 38: 1245–1247 - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
