Elucidating rice cell metabolism under flooding and drought stresses using flux-based modeling and analysis
- PMID: 23753178
- PMCID: PMC3729788
- DOI: 10.1104/pp.113.220178
Elucidating rice cell metabolism under flooding and drought stresses using flux-based modeling and analysis
Abstract
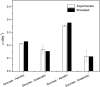
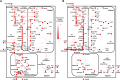
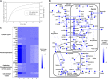
Rice (Oryza sativa) is one of the major food crops in world agriculture, especially in Asia. However, the possibility of subsequent occurrence of flood and drought is a major constraint to its production. Thus, the unique behavior of rice toward flooding and drought stresses has required special attention to understand its metabolic adaptations. However, despite several decades of research investigations, the cellular metabolism of rice remains largely unclear. In this study, in order to elucidate the physiological characteristics in response to such abiotic stresses, we reconstructed what is to our knowledge the first metabolic/regulatory network model of rice, representing two tissue types: germinating seeds and photorespiring leaves. The phenotypic behavior and metabolic states simulated by the model are highly consistent with our suspension culture experiments as well as previous reports. The in silico simulation results of seed-derived rice cells indicated (1) the characteristic metabolic utilization of glycolysis and ethanolic fermentation based on oxygen availability and (2) the efficient sucrose breakdown through sucrose synthase instead of invertase. Similarly, flux analysis on photorespiring leaf cells elucidated the crucial role of plastid-cytosol and mitochondrion-cytosol malate transporters in recycling the ammonia liberated during photorespiration and in exporting the excess redox cofactors, respectively. The model simulations also unraveled the essential role of mitochondrial respiration during drought stress. In the future, the combination of experimental and in silico analyses can serve as a promising approach to understand the complex metabolism of rice and potentially help in identifying engineering targets for improving its productivity as well as enabling stress tolerance.
Figures

Similar articles
-
Ectopic expression of PgRab7 in rice plants (Oryza sativa L.) results in differential tolerance at the vegetative and seed setting stage during salinity and drought stress.Protoplasma. 2017 Jan;254(1):109-124. doi: 10.1007/s00709-015-0914-2. Epub 2015 Dec 14. Protoplasma. 2017. PMID: 26666551
-
From QTL to variety-harnessing the benefits of QTLs for drought, flood and salt tolerance in mega rice varieties of India through a multi-institutional network.Plant Sci. 2016 Jan;242:278-287. doi: 10.1016/j.plantsci.2015.08.008. Epub 2015 Aug 20. Plant Sci. 2016. PMID: 26566845
-
Proteomic dissection of the similar and different responses of wheat to drought, salinity and submergence during seed germination.J Proteomics. 2020 May 30;220:103756. doi: 10.1016/j.jprot.2020.103756. Epub 2020 Mar 20. J Proteomics. 2020. PMID: 32201361
-
Mechanisms of Maturation and Germination in Crop Seeds Exposed to Environmental Stresses with a Focus on Nutrients, Water Status, and Reactive Oxygen Species.Adv Exp Med Biol. 2018;1081:233-257. doi: 10.1007/978-981-13-1244-1_13. Adv Exp Med Biol. 2018. PMID: 30288713 Review.
-
Progress studies of drought-responsive genes in rice.Plant Cell Rep. 2011 Mar;30(3):297-310. doi: 10.1007/s00299-010-0956-z. Epub 2010 Dec 4. Plant Cell Rep. 2011. PMID: 21132431 Review.
Cited by
-
Antioxidant Green Factories: Toward Sustainable Production of Vitamin E in Plant In Vitro Cultures.ACS Omega. 2023 Jan 13;8(4):3586-3605. doi: 10.1021/acsomega.2c05819. eCollection 2023 Jan 31. ACS Omega. 2023. PMID: 36743063 Free PMC article. Review.
-
Plant metabolic modeling: achieving new insight into metabolism and metabolic engineering.Plant Cell. 2014 Oct;26(10):3847-66. doi: 10.1105/tpc.114.130328. Epub 2014 Oct 24. Plant Cell. 2014. PMID: 25344492 Free PMC article. Review.
-
Metabolic and transcriptional regulatory mechanisms underlying the anoxic adaptation of rice coleoptile.AoB Plants. 2014 Jun 3;6:plu026. doi: 10.1093/aobpla/plu026. AoB Plants. 2014. PMID: 24894389 Free PMC article.
-
Hydrogen sulfide enhances nitric oxide-induced tolerance of hypoxia in maize (Zea mays L.).Plant Cell Rep. 2016 Nov;35(11):2325-2340. doi: 10.1007/s00299-016-2037-4. Epub 2016 Aug 11. Plant Cell Rep. 2016. PMID: 27516180
-
SNPeffect: identifying functional roles of SNPs using metabolic networks.Plant J. 2020 Jul;103(2):512-531. doi: 10.1111/tpj.14746. Epub 2020 Apr 18. Plant J. 2020. PMID: 32167625 Free PMC article.
References
-
- Amino SI, Tazawa M. (1988) Uptake and utilization of sugars in cultured rice cells. Plant Cell Physiol 29: 483–487
-
- Atwell BJ, Waters I, Greenway H. (1982) The effect of oxygen and turbulence on elongation of coleoptiles of submergence-tolerant and submergence-intolerant rice cultivars. J Exp Bot 33: 1030–1044
-
- Bailey-Serres J, Voesenek LA. (2008) Flooding stress: acclimations and genetic diversity. Annu Rev Plant Biol 59: 313–339 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

