Genome-wide characterization of transcriptional patterns in high and low antibody responders to rubella vaccination
- PMID: 23658707
- PMCID: PMC3641062
- DOI: 10.1371/journal.pone.0062149
Genome-wide characterization of transcriptional patterns in high and low antibody responders to rubella vaccination
Abstract
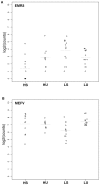
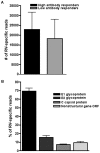
Immune responses to current rubella vaccines demonstrate significant inter-individual variability. We performed mRNA-Seq profiling on PBMCs from high and low antibody responders to rubella vaccination to delineate transcriptional differences upon viral stimulation. Generalized linear models were used to assess the per gene fold change (FC) for stimulated versus unstimulated samples or the interaction between outcome and stimulation. Model results were evaluated by both FC and p-value. Pathway analysis and self-contained gene set tests were performed for assessment of gene group effects. Of 17,566 detected genes, we identified 1,080 highly significant differentially expressed genes upon viral stimulation (p<1.00E(-15), FDR<1.00E(-14)), including various immune function and inflammation-related genes, genes involved in cell signaling, cell regulation and transcription, and genes with unknown function. Analysis by immune outcome and stimulation status identified 27 genes (p≤0.0006 and FDR≤0.30) that responded differently to viral stimulation in high vs. low antibody responders, including major histocompatibility complex (MHC) class I genes (HLA-A, HLA-B and B2M with p = 0.0001, p = 0.0005 and p = 0.0002, respectively), and two genes related to innate immunity and inflammation (EMR3 and MEFV with p = 1.46E(-08) and p = 0.0004, respectively). Pathway and gene set analysis also revealed transcriptional differences in antigen presentation and innate/inflammatory gene sets and pathways between high and low responders. Using mRNA-Seq genome-wide transcriptional profiling, we identified antigen presentation and innate/inflammatory genes that may assist in explaining rubella vaccine-induced immune response variations. Such information may provide new scientific insights into vaccine-induced immunity useful in rational vaccine development and immune response monitoring.
Conflict of interest statement
Figures
Similar articles
-
Transcriptional signatures associated with rubella virus-specific humoral immunity after a third dose of MMR vaccine in women of childbearing age.Eur J Immunol. 2021 Jul;51(7):1824-1838. doi: 10.1002/eji.202049054. Epub 2021 Apr 19. Eur J Immunol. 2021. PMID: 33818775 Free PMC article.
-
Associations between single nucleotide polymorphisms in cellular viral receptors and attachment factor-related genes and humoral immunity to rubella vaccination.PLoS One. 2014 Jun 19;9(6):e99997. doi: 10.1371/journal.pone.0099997. eCollection 2014. PLoS One. 2014. PMID: 24945853 Free PMC article.
-
Polymorphisms in HLA-DPB1 are associated with differences in rubella virus-specific humoral immunity after vaccination.J Infect Dis. 2015 Mar 15;211(6):898-905. doi: 10.1093/infdis/jiu553. Epub 2014 Oct 6. J Infect Dis. 2015. PMID: 25293367 Free PMC article.
-
Rubella virus-associated chronic inflammation in primary immunodeficiency diseases.Curr Opin Allergy Clin Immunol. 2020 Dec;20(6):574-581. doi: 10.1097/ACI.0000000000000694. Curr Opin Allergy Clin Immunol. 2020. PMID: 33044342 Free PMC article. Review.
-
Cutaneous granulomas associated with rubella virus: A clinical review.J Am Acad Dermatol. 2024 Jan;90(1):111-121. doi: 10.1016/j.jaad.2023.05.058. Epub 2023 Jun 2. J Am Acad Dermatol. 2024. PMID: 37271455 Review.
Cited by
-
Comparative Systems Analyses Reveal Molecular Signatures of Clinically tested Vaccine Adjuvants.Sci Rep. 2016 Dec 13;6:39097. doi: 10.1038/srep39097. Sci Rep. 2016. PMID: 27958370 Free PMC article.
-
Gene signatures associated with adaptive humoral immunity following seasonal influenza A/H1N1 vaccination.Genes Immun. 2016 Dec;17(7):371-379. doi: 10.1038/gene.2016.34. Epub 2016 Aug 18. Genes Immun. 2016. PMID: 27534615 Free PMC article.
-
Transcriptomic signatures induced by the Ebola virus vaccine rVSVΔG-ZEBOV-GP in adult cohorts in Europe, Africa, and North America: a molecular biomarker study.Lancet Microbe. 2022 Feb;3(2):e113-e123. doi: 10.1016/S2666-5247(21)00235-4. Epub 2021 Dec 6. Lancet Microbe. 2022. PMID: 35544042 Free PMC article.
-
System-Wide Associations between DNA-Methylation, Gene Expression, and Humoral Immune Response to Influenza Vaccination.PLoS One. 2016 Mar 31;11(3):e0152034. doi: 10.1371/journal.pone.0152034. eCollection 2016. PLoS One. 2016. PMID: 27031986 Free PMC article.
-
Transcriptome Analysis of Peripheral Blood Mononuclear Cells in SARS-CoV-2 Naïve and Recovered Individuals Vaccinated With Inactivated Vaccine.Front Cell Infect Microbiol. 2022 Feb 3;11:821828. doi: 10.3389/fcimb.2021.821828. eCollection 2021. Front Cell Infect Microbiol. 2022. PMID: 35186784 Free PMC article.
References
-
- Hill AV (2001) The genomics and genetics of human infectious disease susceptibility. Annu Rev Genomics Hum Genet 2: 373–400. - PubMed
-
- Hill AV (2006) Aspects of genetic susceptibility to human infectious diseases. Annu Rev Genet 40: 469–486. - PubMed
-
- Poland GA, Ovsyannikova IG, Jacobson RM, Smith DI (2007) Heterogeneity in vaccine immune response: the role of immunogenetics and the emerging field of vaccinomics. Clin Pharmacol Ther 82: 653–664 6100415 [pii];10.1038/sj.clpt.6100415 [doi]. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials
Miscellaneous

