Specific amino acid substitutions in the S protein prevent its excretion in vitro and may contribute to occult hepatitis B virus infection
- PMID: 23658444
- PMCID: PMC3700177
- DOI: 10.1128/JVI.00710-13
Specific amino acid substitutions in the S protein prevent its excretion in vitro and may contribute to occult hepatitis B virus infection
Abstract
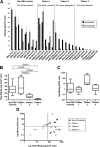
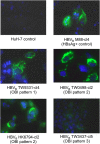
Occult hepatitis B virus (HBV) infection (OBI) is defined as low plasma level of HBV DNA with undetectable HBV surface antigen (HBsAg) outside the preseroconversion window period. The mechanisms leading to OBI remain largely unknown. The potential role of specific amino acid substitutions in the S protein from OBI in HBsAg production and excretion was examined in vitro. HBsAg was quantified in culture supernatants and cell extracts of HuH-7 cells transiently transfected with plasmids containing the S gene of eight HBsAg(+) controls and 18 OBI clones. The intracellular (IC)/extracellular (EC) HBsAg production ratio was ∼1.0 for the majority of controls. Three IC/EC HBsAg patterns were observed in OBI strains clones: pattern 1, an IC/EC ratio of 1.0, was found in 5/18 OBI clones, pattern 2, detectable IC but low or undetectable EC HBsAg (IC/EC, 7.0 to 800), was found in 6/18 OBIs, and pattern 3, low or undetectable IC and EC HBsAg, was found in 7/18 clones. Intracellular immunofluorescence staining showed that in pattern 2, HBsAg was concentrated around the nucleus, suggesting retention in the endoplasmic reticulum. The substitution M75T, Y100S, or P178R was present in 4/6 pattern 2 OBI clones. Site-directed-mutagenesis-corrected mutations reversed HBsAg excretion to pattern 1 and, when introduced into a control clone, induced pattern 2 except for Y100S. In a control and several OBIs, variants of a given quasispecies expressed HBsAg according to different patterns. However, the P178R substitution present in all cloned sequences of two OBI strains may contribute significantly to the OBI phenotype.
Figures

Similar articles
-
Role of core protein mutations in the development of occult HBV infection.J Hepatol. 2021 Jun;74(6):1303-1314. doi: 10.1016/j.jhep.2020.12.023. Epub 2021 Jan 13. J Hepatol. 2021. PMID: 33453326
-
T cell responses and viral variability in blood donation candidates with occult hepatitis B infection.J Hepatol. 2012 Apr;56(4):765-74. doi: 10.1016/j.jhep.2011.11.011. Epub 2011 Dec 13. J Hepatol. 2012. PMID: 22173156
-
M133S mutation possibly involve in the ER stress and mitophagy pathway in maintenance hemodialysis patients with occult hepatitis B infection.Sci Rep. 2024 Jun 17;14(1):13981. doi: 10.1038/s41598-024-64943-3. Sci Rep. 2024. PMID: 38886481 Free PMC article.
-
Occult hepatitis B virus infection: detection and significance.Dig Dis. 2010;28(1):116-25. doi: 10.1159/000282074. Epub 2010 May 7. Dig Dis. 2010. PMID: 20460899 Review.
-
Occult hepatitis B virus infection: implications in transfusion.Vox Sang. 2004 Feb;86(2):83-91. doi: 10.1111/j.0042-9007.2004.00406.x. Vox Sang. 2004. PMID: 15023176 Review.
Cited by
-
Decreasing prevalence of Hepatitis B and absence of Hepatitis C Virus infection in the Warao indigenous population of Venezuela.PLoS One. 2018 May 25;13(5):e0197662. doi: 10.1371/journal.pone.0197662. eCollection 2018. PLoS One. 2018. PMID: 29799873 Free PMC article.
-
Molecular mechanisms underlying HBsAg negativity in occult HBV infection.Eur J Clin Microbiol Infect Dis. 2015 Sep;34(9):1709-31. doi: 10.1007/s10096-015-2422-x. Epub 2015 Jun 24. Eur J Clin Microbiol Infect Dis. 2015. PMID: 26105620 Review.
-
Functional analysis of 'a' determinant mutations associated with occult HBV in HIV-positive South Africans.J Gen Virol. 2016 Jul;97(7):1615-1624. doi: 10.1099/jgv.0.000469. Epub 2016 Mar 31. J Gen Virol. 2016. PMID: 27031988 Free PMC article.
-
Recent and occult hepatitis B virus infections among blood donors in the United States.Transfusion. 2019 Feb;59(2):601-611. doi: 10.1111/trf.15057. Epub 2018 Nov 30. Transfusion. 2019. PMID: 30499591 Free PMC article.
-
Investigation of a Novel Hepatitis B Virus Surface Antigen (HBsAg) Escape Mutant Affecting Immunogenicity.PLoS One. 2017 Jan 3;12(1):e0167871. doi: 10.1371/journal.pone.0167871. eCollection 2017. PLoS One. 2017. PMID: 28045894 Free PMC article.
References
-
- Raimondo G, Allain JP, Brunetto MR, Buendia MA, Chen DS, Colombo M, Craxi A, Donato F, Ferrari C, Gaeta GB, Gerlich WH, Levrero M, Locarnini S, Michalak T, Mondelli MU, Pawlotsky JM, Pollicino T, Prati D, Puoti M, Samuel D, Shouval D, Smedile A, Squadrito G, Trépo C, Villa E, Will H, Zanetti AR, Zoulim F. 2008. Statements from the Taormina expert meeting on occult hepatitis B virus infection. J. Hepatol. 49:652–657 - PubMed
-
- Raimondo G, Pollicino T, Romano L, Zanetti AR. 2010. A 2010 update on occult hepatitis B infection. Pathol. Biol. 58:254–257 - PubMed
-
- Pollicino T, Raffa G, Costantino L, Lisa A, Campello C, Squadrito G, Levrero M, Raimondo G. 2007. Molecular and functional analysis of occult hepatitis B virus isolates from patients with hepatocellular carcinoma. Hepatology 45:277–285 - PubMed
-
- Candotti D, Grabarczyk P, Ghiazza P, Roig R, Casamitjana N, Iudicone P, Schmidt M, Bird A, Crookes R, Brojer E, Miceli M, Amiri A, Li C, Allain JP. 2008. Characterization of occult hepatitis B virus from blood donors carrying genotype A2 or genotype D strains. J. Hepatol. 49:537–547 - PubMed
-
- Huang CH, Yuan Q, Chen PJ, Zhang YL, Chen CR, Zheng QB, Yeh SH, Yu H, Xue Y, Chen YX, Liu PG, Ge SX, Zhang J, Xia NS. 2012. Influence of mutations in hepatitis B virus surface protein on viral antigenicity and phenotype in occult HBV strains from blood donors. J. Hepatol. 57:720–729 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

