Epigenetic silencing of the proapoptotic gene BIM in anaplastic large cell lymphoma through an MeCP2/SIN3a deacetylating complex
- PMID: 23633923
- PMCID: PMC3638354
- DOI: 10.1593/neo.121784
Epigenetic silencing of the proapoptotic gene BIM in anaplastic large cell lymphoma through an MeCP2/SIN3a deacetylating complex
Abstract
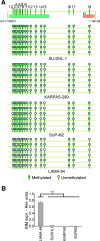
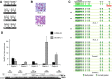
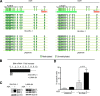
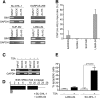
BIM is a proapoptotic member of the Bcl-2 family. Here, we investigated the epigenetic status of the BIM locus in NPM/ALK+ anaplastic large cell lymphoma (ALCL) cell lines and in lymph node biopsies from NPM/ALK+ ALCL patients. We show that BIM is epigenetically silenced in cell lines and lymph node specimens and that treatment with the deacetylase inhibitor trichostatin A restores the histone acetylation, strongly upregulates BIM expression, and induces cell death. BIM silencing occurs through recruitment of MeCP2 and the SIN3a/histone deacetylase 1/2 (HDAC1/2) corepressor complex. This event requires BIM CpG methylation/demethylation with 5-azacytidine that leads to detachment of the MeCP2 corepressor complex and reacetylation of the histone tails. Treatment with the ALK inhibitor PF2341066 or with an inducible shRNA targeting NPM/ALK does not restore BIM locus reacetylation; however, enforced expression of NPM/ALK in an NPM/ALK-negative cell line significantly increases the methylation at the BIM locus. This study demonstrates that BIM is epigenetically silenced in NPM/ALK-positive cells through recruitment of the SIN3a/HDAC1/2 corepressor complex and that NPM/ALK is dispensable to maintain BIM epigenetic silencing but is able to act as an inducer of BIM methylation.
Figures

Similar articles
-
Epstein-barr virus latency in B cells leads to epigenetic repression and CpG methylation of the tumour suppressor gene Bim.PLoS Pathog. 2009 Jun;5(6):e1000492. doi: 10.1371/journal.ppat.1000492. Epub 2009 Jun 26. PLoS Pathog. 2009. PMID: 19557159 Free PMC article.
-
IGF-1 suppresses Bim expression in multiple myeloma via epigenetic and posttranslational mechanisms.Blood. 2010 Mar 25;115(12):2430-40. doi: 10.1182/blood-2009-07-232801. Epub 2010 Jan 19. Blood. 2010. PMID: 20086250
-
The heat shock protein-90 co-chaperone, Cyclophilin 40, promotes ALK-positive, anaplastic large cell lymphoma viability and its expression is regulated by the NPM-ALK oncoprotein.BMC Cancer. 2012 Jun 8;12:229. doi: 10.1186/1471-2407-12-229. BMC Cancer. 2012. PMID: 22681779 Free PMC article.
-
Nucleophosmin-anaplastic lymphoma kinase: the ultimate oncogene and therapeutic target.Blood. 2017 Feb 16;129(7):823-831. doi: 10.1182/blood-2016-05-717793. Epub 2016 Nov 22. Blood. 2017. PMID: 27879258 Review.
-
The t(2;5) in human lymphomas.Leuk Lymphoma. 1998 Apr;29(3-4):249-56. doi: 10.3109/10428199809068562. Leuk Lymphoma. 1998. PMID: 9684923 Review.
Cited by
-
Mecp2 promotes the anti-inflammatory effect of alpinetin via epigenetic modification crosstalk.J Cell Mol Med. 2024 Jul;28(13):e18510. doi: 10.1111/jcmm.18510. J Cell Mol Med. 2024. PMID: 38953409 Free PMC article.
-
SETBP1 induces transcription of a network of development genes by acting as an epigenetic hub.Nat Commun. 2018 Jun 6;9(1):2192. doi: 10.1038/s41467-018-04462-8. Nat Commun. 2018. PMID: 29875417 Free PMC article.
-
Redundant and nonredundant roles for Cdc42 and Rac1 in lymphomas developed in NPM-ALK transgenic mice.Blood. 2016 Mar 10;127(10):1297-306. doi: 10.1182/blood-2015-11-683052. Epub 2016 Jan 8. Blood. 2016. PMID: 26747246 Free PMC article.
-
Cancer subclonal genetic architecture as a key to personalized medicine.Neoplasia. 2013 Dec;15(12):1410-20. doi: 10.1593/neo.131972. Neoplasia. 2013. PMID: 24403863 Free PMC article.
-
EBV and Apoptosis: The Viral Master Regulator of Cell Fate?Viruses. 2017 Nov 13;9(11):339. doi: 10.3390/v9110339. Viruses. 2017. PMID: 29137176 Free PMC article. Review.
References
-
- Fletcher JI, Huang DC. Controlling the cell death mediators Bax and Bak: puzzles and conundrums. Cell Cycle. 2008;7:39–44. E-pub 2007 Oct 2016. - PubMed
-
- Bouillet P, Metcalf D, Huang DC, Tarlinton DM, Kay TW, Kontgen F, Adams JM, Strasser A. Proapoptotic Bcl-2 relative Bim required for certain apoptotic responses, leukocyte homeostasis, and to preclude autoimmunity. Science. 1999;286:1735–1738. - PubMed
-
- Tagawa H, Karnan S, Suzuki R, Matsuo K, Zhang X, Ota A, Morishima Y, Nakamura S, Seto M. Genome-wide array-based CGH for mantle cell lymphoma: identification of homozygous deletions of the proapoptotic gene BIM. Oncogene. 2005;24:1348–1358. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous
