Systems scale interactive exploration reveals quantitative and qualitative differences in response to influenza and pneumococcal vaccines
- PMID: 23601689
- PMCID: PMC3681204
- DOI: 10.1016/j.immuni.2012.12.008
Systems scale interactive exploration reveals quantitative and qualitative differences in response to influenza and pneumococcal vaccines
Abstract
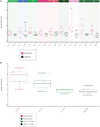
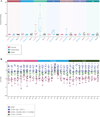
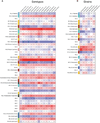
Systems immunology approaches were employed to investigate innate and adaptive immune responses to influenza and pneumococcal vaccines. These two non-live vaccines show different magnitudes of transcriptional responses at different time points after vaccination. Software solutions were developed to explore correlates of vaccine efficacy measured as antibody titers at day 28. These enabled a further dissection of transcriptional responses. Thus, the innate response, measured within hours in the peripheral blood, was dominated by an interferon transcriptional signature after influenza vaccination and by an inflammation signature after pneumococcal vaccination. Day 7 plasmablast responses induced by both vaccines was more pronounced after pneumococcal vaccination. Together, these results suggest that comparing global immune responses elicited by different vaccines will be critical to our understanding of the immune mechanisms underpinning successful vaccination.
Copyright © 2013 Elsevier Inc. All rights reserved.
Figures
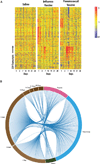
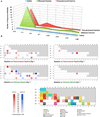
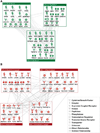
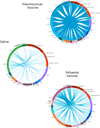
Comment in
-
From finger pricks to point-and-click.Immunity. 2013 Apr 18;38(4):622-4. doi: 10.1016/j.immuni.2013.04.003. Immunity. 2013. PMID: 23601677 Free PMC article.
Similar articles
-
Enhanced protective responses to a serotype-independent pneumococcal vaccine when combined with an inactivated influenza vaccine.Clin Sci (Lond). 2017 Jan 1;131(2):169-180. doi: 10.1042/CS20160475. Epub 2016 Nov 24. Clin Sci (Lond). 2017. PMID: 27885052
-
The response to vaccination against influenza A(H1N1) 2009, seasonal influenza and Streptococcus pneumoniae in adult outpatients with ongoing treatment for cancer with and without rituximab.Acta Oncol. 2014 Sep;53(9):1212-20. doi: 10.3109/0284186X.2014.914243. Epub 2014 May 28. Acta Oncol. 2014. PMID: 24865118
-
[Characteristics and Differences in Pneumococcal Vaccines: 1. Polysaccharide vaccine].Nihon Naika Gakkai Zasshi. 2015 Nov 10;104(11):2314-23. doi: 10.2169/naika.104.2314. Nihon Naika Gakkai Zasshi. 2015. PMID: 28520389 Japanese. No abstract available.
-
[Evidence and guidelines for influenza and pneumococcal vaccines for the elderly].Ugeskr Laeger. 2013 Nov 18;175(47):2861-4. Ugeskr Laeger. 2013. PMID: 24629386 Review. Danish.
-
Vaccine responsiveness in the elderly: best practice for the clinic.Expert Rev Vaccines. 2014 Jul;13(7):885-94. doi: 10.1586/14760584.2014.924403. Epub 2014 Jun 3. Expert Rev Vaccines. 2014. PMID: 24890875 Review.
Cited by
-
Aging-dependent alterations in gene expression and a mitochondrial signature of responsiveness to human influenza vaccination.Aging (Albany NY). 2015 Jan;7(1):38-52. doi: 10.18632/aging.100720. Aging (Albany NY). 2015. PMID: 25596819 Free PMC article.
-
Metabolomic and transcriptomic signatures of influenza vaccine response in healthy young and older adults.Aging Cell. 2022 Sep;21(9):e13682. doi: 10.1111/acel.13682. Epub 2022 Aug 23. Aging Cell. 2022. PMID: 35996998 Free PMC article.
-
Gene Expression Signatures Associated With Immune and Virological Responses to Therapeutic Vaccination With Dendritic Cells in HIV-Infected Individuals.Front Immunol. 2019 Apr 24;10:874. doi: 10.3389/fimmu.2019.00874. eCollection 2019. Front Immunol. 2019. PMID: 31105698 Free PMC article. Clinical Trial.
-
Modular transcriptional repertoire analyses of adults with systemic lupus erythematosus reveal distinct type I and type II interferon signatures.Arthritis Rheumatol. 2014 Jun;66(6):1583-95. doi: 10.1002/art.38628. Arthritis Rheumatol. 2014. PMID: 24644022 Free PMC article.
-
Immunosenescence: A systems-level overview of immune cell biology and strategies for improving vaccine responses.Exp Gerontol. 2019 Sep;124:110632. doi: 10.1016/j.exger.2019.110632. Epub 2019 Jun 13. Exp Gerontol. 2019. PMID: 31201918 Free PMC article.
References
-
- Alakulppi N, Seikku P, Jaatinen T, Holmberg C, Laine J. Feasibility of diagnosing subclinical renal allograft rejection in children by whole blood gene expression analysis. Transplantation. 2008;86:1222–1228. - PubMed
-
- Benjamini Y, Hochberg Y. Controlling the False Discovery Rate: A Practical and Powerful Approach to Multiple Testing. J.R. Stat. Soc. 1995;57:289–300.
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases

