Epstein-Barr virus maintains lymphomas via its miRNAs
- PMID: 23503461
- PMCID: PMC3690170
- DOI: 10.1038/onc.2013.71
Epstein-Barr virus maintains lymphomas via its miRNAs
Abstract
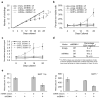
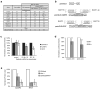
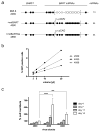
Epstein-Barr virus (EBV) has evolved exquisite controls over its host cells, human B lymphocytes, not only directing these cells during latency to proliferate and thereby expand the pool of infected cells, but also to survive and thereby persist for the lifetime of the infected individual. Although these activities ensure the virus is successful, they also make the virus oncogenic, particularly when infected people are immunosuppressed. Here we show, strikingly, that one set of EBV's microRNAs (miRNAs) both sustain Burkitt's lymphoma (BL) cells in the absence of other viral oncogenes and promote the transformation of primary B lymphocytes. BL cells were engineered to lose EBV and found to die by apoptosis and could be rescued by constitutively expressing viral miRNAs in them. Two of these EBV miRNAs were found to target caspase 3 to inhibit apoptosis at physiological concentrations.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
Similar articles
-
Epstein-Barr virus provides a survival factor to Burkitt's lymphomas.Proc Natl Acad Sci U S A. 2003 Nov 25;100(24):14269-74. doi: 10.1073/pnas.2336099100. Epub 2003 Nov 5. Proc Natl Acad Sci U S A. 2003. PMID: 14603034 Free PMC article.
-
Heterogeneous expression of Epstein-Barr virus latent proteins in endemic Burkitt's lymphoma.Blood. 1995 Jul 15;86(2):659-65. Blood. 1995. PMID: 7605996
-
Three restricted forms of Epstein-Barr virus latency counteracting apoptosis in c-myc-expressing Burkitt lymphoma cells.Proc Natl Acad Sci U S A. 2006 Oct 3;103(40):14935-40. doi: 10.1073/pnas.0509988103. Epub 2006 Sep 25. Proc Natl Acad Sci U S A. 2006. PMID: 17001014 Free PMC article.
-
Epstein-Barr virus-encoded microRNAs as regulators in host immune responses.Int J Biol Sci. 2018 Apr 5;14(5):565-576. doi: 10.7150/ijbs.24562. eCollection 2018. Int J Biol Sci. 2018. PMID: 29805308 Free PMC article. Review.
-
EBV regulates c-MYC, apoptosis, and tumorigenicity in Burkitt's lymphoma.Curr Top Microbiol Immunol. 2001;258:153-60. doi: 10.1007/978-3-642-56515-1_10. Curr Top Microbiol Immunol. 2001. PMID: 11443860 Review. No abstract available.
Cited by
-
The presence of Epstein-Barr virus significantly impacts the transcriptional profile in immunodeficiency-associated Burkitt lymphoma.Front Microbiol. 2015 Jun 10;6:556. doi: 10.3389/fmicb.2015.00556. eCollection 2015. Front Microbiol. 2015. PMID: 26113842 Free PMC article.
-
EBV BART MicroRNAs Target Multiple Pro-apoptotic Cellular Genes to Promote Epithelial Cell Survival.PLoS Pathog. 2015 Jun 12;11(6):e1004979. doi: 10.1371/journal.ppat.1004979. eCollection 2015 Jun. PLoS Pathog. 2015. PMID: 26070070 Free PMC article.
-
Epstein-Barr virus and Burkitt lymphoma.Chin J Cancer. 2014 Dec;33(12):609-19. doi: 10.5732/cjc.014.10190. Epub 2014 Nov 21. Chin J Cancer. 2014. PMID: 25418195 Free PMC article. Review.
-
EBV Persistence--Introducing the Virus.Curr Top Microbiol Immunol. 2015;390(Pt 1):151-209. doi: 10.1007/978-3-319-22822-8_8. Curr Top Microbiol Immunol. 2015. PMID: 26424647 Free PMC article. Review.
-
Divergent viral presentation among human tumors and adjacent normal tissues.Sci Rep. 2016 Jun 24;6:28294. doi: 10.1038/srep28294. Sci Rep. 2016. PMID: 27339696 Free PMC article.
References
-
- Vereide D, Sugden B. Insights into the evolution of lymphomas induced by Epstein-Barr virus. Advances in cancer research. 2010;108:1–19. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

