Cytoplasmic and nuclear quality control and turnover of single-stranded RNA modulate post-transcriptional gene silencing in plants
- PMID: 23482394
- PMCID: PMC3632135
- DOI: 10.1093/nar/gkt152
Cytoplasmic and nuclear quality control and turnover of single-stranded RNA modulate post-transcriptional gene silencing in plants
Abstract
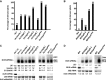
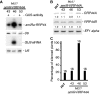
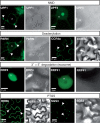
Eukaryotic RNA quality control (RQC) uses both endonucleolytic and exonucleolytic degradation to eliminate dysfunctional RNAs. In addition, endogenous and exogenous RNAs are degraded through post-transcriptional gene silencing (PTGS), which is triggered by the production of double-stranded (ds)RNAs and proceeds through short-interfering (si)RNA-directed ARGONAUTE-mediated endonucleolytic cleavage. Compromising cytoplasmic or nuclear 5'-3' exoribonuclease function enhances sense-transgene (S)-PTGS in Arabidopsis, suggesting that these pathways compete for similar RNA substrates. Here, we show that impairing nonsense-mediated decay, deadenylation or exosome activity enhanced S-PTGS, which requires host RNA-dependent RNA polymerase 6 (RDR6/SGS2/SDE1) and SUPPRESSOR OF GENE SILENCING 3 (SGS3) for the transformation of single-stranded RNA into dsRNA to trigger PTGS. However, these RQC mutations had no effect on inverted-repeat-PTGS, which directly produces hairpin dsRNA through transcription. Moreover, we show that these RQC factors are nuclear and cytoplasmic and are found in two RNA degradation foci in the cytoplasm: siRNA-bodies and processing-bodies. We propose a model of single-stranded RNA tug-of-war between RQC and S-PTGS that ensures the correct partitioning of RNA substrates among these RNA degradation pathways.
Figures


Similar articles
-
The Nuclear Ribonucleoprotein SmD1 Interplays with Splicing, RNA Quality Control, and Posttranscriptional Gene Silencing in Arabidopsis.Plant Cell. 2016 Feb;28(2):426-38. doi: 10.1105/tpc.15.01045. Epub 2016 Feb 3. Plant Cell. 2016. PMID: 26842463 Free PMC article.
-
In plants, decapping prevents RDR6-dependent production of small interfering RNAs from endogenous mRNAs.Nucleic Acids Res. 2015 Mar 11;43(5):2902-13. doi: 10.1093/nar/gkv119. Epub 2015 Feb 18. Nucleic Acids Res. 2015. PMID: 25694514 Free PMC article.
-
SUPERKILLER Complex Components Are Required for the RNA Exosome-Mediated Control of Cuticular Wax Biosynthesis in Arabidopsis Inflorescence Stems.Plant Physiol. 2016 Jun;171(2):960-73. doi: 10.1104/pp.16.00450. Epub 2016 Apr 28. Plant Physiol. 2016. PMID: 27208312 Free PMC article.
-
RNA Quality Control as a Key to Suppressing RNA Silencing of Endogenous Genes in Plants.Mol Plant. 2016 Jun 6;9(6):826-36. doi: 10.1016/j.molp.2016.03.011. Epub 2016 Mar 30. Mol Plant. 2016. PMID: 27045817 Free PMC article. Review.
-
The diversity of post-transcriptional gene silencing mediated by small silencing RNAs in plants.Essays Biochem. 2020 Dec 7;64(6):919-930. doi: 10.1042/EBC20200006. Essays Biochem. 2020. PMID: 32885814 Review.
Cited by
-
Innate, translation-dependent silencing of an invasive transposon in Arabidopsis.EMBO Rep. 2022 Feb 3;23(3):e53400. doi: 10.15252/embr.202153400. Epub 2021 Dec 21. EMBO Rep. 2022. PMID: 34931432 Free PMC article.
-
Catalytic activities, molecular connections, and biological functions of plant RNA exosome complexes.Plant Cell. 2022 Mar 4;34(3):967-988. doi: 10.1093/plcell/koab310. Plant Cell. 2022. PMID: 34954803 Free PMC article. Review.
-
The Zinc-Finger Protein SOP1 Is Required for a Subset of the Nuclear Exosome Functions in Arabidopsis.PLoS Genet. 2016 Feb 1;12(2):e1005817. doi: 10.1371/journal.pgen.1005817. eCollection 2016 Feb. PLoS Genet. 2016. PMID: 26828932 Free PMC article.
-
The Nuclear Ribonucleoprotein SmD1 Interplays with Splicing, RNA Quality Control, and Posttranscriptional Gene Silencing in Arabidopsis.Plant Cell. 2016 Feb;28(2):426-38. doi: 10.1105/tpc.15.01045. Epub 2016 Feb 3. Plant Cell. 2016. PMID: 26842463 Free PMC article.
-
Contrasting epigenetic control of transgenes and endogenous genes promotes post-transcriptional transgene silencing in Arabidopsis.Nat Commun. 2021 May 13;12(1):2787. doi: 10.1038/s41467-021-22995-3. Nat Commun. 2021. PMID: 33986281 Free PMC article.
References
-
- Staiger D, Korneli C, Lummer M, Navarro L. Emerging role for RNA-based regulation in plant immunity. New Phytol. 2012;197:394–404. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

