Complete cardiac regeneration in a mouse model of myocardial infarction
- PMID: 23425860
- PMCID: PMC3615162
- DOI: 10.18632/aging.100526
Complete cardiac regeneration in a mouse model of myocardial infarction
Abstract
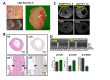
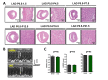
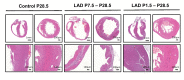
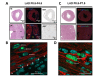
Cardiac remodeling and subsequent heart failure remain critical issues after myocardial infarction despite improved treatment and reperfusion strategies. Recently, complete cardiac regeneration has been demonstrated in fish and newborn mice following resection of the cardiac apex. However, it remained entirely unclear whether the mammalian heart can also completely regenerate following a complex cardiac ischemic injury. We established a protocol to induce a severe heart attack in one-day-old mice using left anterior descending artery (LAD) ligation. LAD ligation triggered substantial cardiac injury in the left ventricle defined by Caspase 3 activation and massive cell death. Ischemia-induced cardiomyocyte death was also visible on day 4 after LAD ligation. Remarkably, 7 days after the initial ischemic insult, we observed complete cardiac regeneration without any signs of tissue damage or scarring. This tissue regeneration translated into long-term normal heart functions as assessed by echocardiography. In contrast, LAD ligations in 7-day-old mice resulted in extensive scarring comparable to adult mice, indicating that the regenerative capacity for complete cardiac healing after heart attacks can be traced to the first week after birth. RNAseq analyses of hearts on day 1, day 3, and day 10 and comparing LAD-ligated and sham-operated mice surprisingly revealed a transcriptional programme of major changes in genes mediating mitosis and cell division between days 1, 3 and 10 postnatally and a very limited set of genes, including genes regulating cell cycle and extracellular matrix synthesis, being differentially regulated in the regenerating hearts. We present for the first time a mammalian model of complete cardiac regeneration following a severe ischemic cardiac injury. This novel model system provides the unique opportunity to uncover molecular and cellular pathways that can induce cardiac regeneration after ischemic injury, findings that one day could be translated to human heart attack patients.
Conflict of interest statement
The authors of this manuscript have no conflict of interests to declare.
Figures
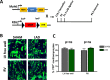
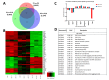

Similar articles
-
A reproducible protocol for neonatal ischemic injury and cardiac regeneration in neonatal mice.Basic Res Cardiol. 2016 Nov;111(6):64. doi: 10.1007/s00395-016-0580-3. Epub 2016 Sep 24. Basic Res Cardiol. 2016. PMID: 27665606 Free PMC article.
-
Therapeutic effect of a novel Wnt pathway inhibitor on cardiac regeneration after myocardial infarction.Clin Sci (Lond). 2017 Dec 8;131(24):2919-2932. doi: 10.1042/CS20171256. Print 2017 Dec 15. Clin Sci (Lond). 2017. PMID: 29162747
-
Regenerative cross talk between cardiac cells and macrophages.Am J Physiol Heart Circ Physiol. 2021 Jun 1;320(6):H2211-H2221. doi: 10.1152/ajpheart.00056.2021. Epub 2021 Mar 26. Am J Physiol Heart Circ Physiol. 2021. PMID: 33769920 Free PMC article.
-
Cell Cycle-Mediated Cardiac Regeneration in the Mouse Heart.Curr Cardiol Rep. 2019 Sep 16;21(10):131. doi: 10.1007/s11886-019-1206-9. Curr Cardiol Rep. 2019. PMID: 31529165 Free PMC article. Review.
-
Adult Cardiomyocyte Proliferation: a New Insight for Myocardial Infarction Therapy.J Cardiovasc Transl Res. 2021 Jun;14(3):457-466. doi: 10.1007/s12265-020-10067-8. Epub 2020 Aug 20. J Cardiovasc Transl Res. 2021. PMID: 32820393 Review.
Cited by
-
Endocrine Influence on Cardiac Metabolism in Development and Regeneration.Endocrinology. 2021 Sep 1;162(9):bqab081. doi: 10.1210/endocr/bqab081. Endocrinology. 2021. PMID: 33880553 Free PMC article. Review.
-
The non-coding road towards cardiac regeneration.J Cardiovasc Transl Res. 2013 Dec;6(6):909-23. doi: 10.1007/s12265-013-9486-8. J Cardiovasc Transl Res. 2013. PMID: 23797382 Review.
-
A Unique Collateral Artery Development Program Promotes Neonatal Heart Regeneration.Cell. 2019 Feb 21;176(5):1128-1142.e18. doi: 10.1016/j.cell.2018.12.023. Epub 2019 Jan 24. Cell. 2019. PMID: 30686582 Free PMC article.
-
Accelerated Growth, Differentiation, and Ploidy with Reduced Proliferation of Right Ventricular Cardiomyocytes in Children with Congenital Heart Defect Tetralogy of Fallot.Cells. 2022 Jan 5;11(1):175. doi: 10.3390/cells11010175. Cells. 2022. PMID: 35011735 Free PMC article.
-
Mechanisms and Therapeutic Targets of Cardiac Regeneration: Closing the Age Gap.Front Cardiovasc Med. 2018 Feb 5;5:7. doi: 10.3389/fcvm.2018.00007. eCollection 2018. Front Cardiovasc Med. 2018. PMID: 29459901 Free PMC article. Review.
References
-
- Yusuf S, Reddy S, Ounpuu S, Anand S. Global burden of cardiovascular diseases: part I: general considerations, the epidemiologic transition, risk factors, and impact of urbanization. Circulation. 2001;104:2746–2753. - PubMed
-
- A.H.A. http://www.americanheart.org/. Available at: http://www.americanheart.org/.
-
- Lloyd-Jones D, Adams R, Carnethon M, De Simone G, Ferguson TB, Flegal K, Ford E, Furie K, Go A, Greenlund K, Haase N, Hailpern S, Ho M, Howard V, Kissela B, Kittner S, et al. Heart disease and stroke statistics--2009 update: a report from the American Heart Association Statistics Committee and Stroke Statistics Subcommittee. Circulation. 2009;119:e21–181. - PubMed
-
- Mudd JO, Kass DA. Tackling heart failure in the twenty-first century. Nature. 2008;451:919–928. - PubMed
-
- Hosoda T, D'Amario D, Cabral-Da-Silva MC, Zheng H, Padin-Iruegas ME, Ogorek B, Ferreira-Martins J, Yasuzawa-Amano S, Amano K, Ide-Iwata N, Cheng W, Rota M, Urbanek K, Kajstura J, Anversa P, Leri A. Clonality of mouse and human cardiomyogenesis in vivo. Proc Natl Acad Sci U S A. 2009;106:17169–17174. - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials
Miscellaneous

