Proteasomal degradation of herpes simplex virus capsids in macrophages releases DNA to the cytosol for recognition by DNA sensors
- PMID: 23345332
- PMCID: PMC3578088
- DOI: 10.4049/jimmunol.1202749
Proteasomal degradation of herpes simplex virus capsids in macrophages releases DNA to the cytosol for recognition by DNA sensors
Abstract
The innate immune system is important for control of infections, including herpesvirus infections. Intracellular DNA potently stimulates antiviral IFN responses. It is known that plasmacytoid dendritic cells sense herpesvirus DNA in endosomes via TLR9 and that nonimmune tissue cells can sense herpesvirus DNA in the nucleus. However, it remains unknown how and where myeloid cells, such as macrophages and conventional dendritic cells, detect infections with herpesviruses. In this study, we demonstrate that the HSV-1 capsid was ubiquitinated in the cytosol and degraded by the proteasome, hence releasing genomic DNA into the cytoplasm for detection by DNA sensors. In this context, the DNA sensor IFN-γ-inducible 16 is important for induction of IFN-β in human macrophages postinfection with HSV-1 and CMV. Viral DNA localized to the same cytoplasmic regions as did IFN-γ-inducible 16, with DNA sensing being independent of viral nuclear entry. Thus, proteasomal degradation of herpesvirus capsids releases DNA to the cytoplasm for recognition by DNA sensors.
Figures
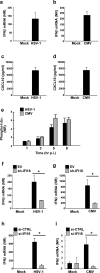
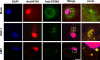
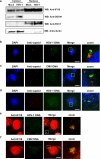
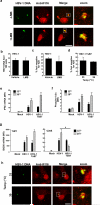
Similar articles
-
Cellular Requirements for Sensing and Elimination of Incoming HSV-1 DNA and Capsids.J Interferon Cytokine Res. 2019 Apr;39(4):191-204. doi: 10.1089/jir.2018.0141. Epub 2019 Mar 11. J Interferon Cytokine Res. 2019. PMID: 30855198
-
Type I interferon production during herpes simplex virus infection is controlled by cell-type-specific viral recognition through Toll-like receptor 9, the mitochondrial antiviral signaling protein pathway, and novel recognition systems.J Virol. 2007 Dec;81(24):13315-24. doi: 10.1128/JVI.01167-07. Epub 2007 Oct 3. J Virol. 2007. PMID: 17913820 Free PMC article.
-
Viral DNA Sensors IFI16 and Cyclic GMP-AMP Synthase Possess Distinct Functions in Regulating Viral Gene Expression, Immune Defenses, and Apoptotic Responses during Herpesvirus Infection.mBio. 2016 Nov 15;7(6):e01553-16. doi: 10.1128/mBio.01553-16. mBio. 2016. PMID: 27935834 Free PMC article.
-
Herpesvirus Genome Recognition Induced Acetylation of Nuclear IFI16 Is Essential for Its Cytoplasmic Translocation, Inflammasome and IFN-β Responses.PLoS Pathog. 2015 Jul 2;11(7):e1005019. doi: 10.1371/journal.ppat.1005019. eCollection 2015 Jul. PLoS Pathog. 2015. PMID: 26134128 Free PMC article.
-
The emerging role of nuclear viral DNA sensors.J Biol Chem. 2015 Oct 30;290(44):26412-21. doi: 10.1074/jbc.R115.652289. Epub 2015 Sep 9. J Biol Chem. 2015. PMID: 26354430 Free PMC article. Review.
Cited by
-
Innate immune recognition of DNA: A recent history.Virology. 2015 May;479-480:146-52. doi: 10.1016/j.virol.2015.03.013. Epub 2015 Mar 26. Virology. 2015. PMID: 25816762 Free PMC article. Review.
-
Cytokines and chemokines profile in encephalitis patients: A meta-analysis.PLoS One. 2022 Sep 1;17(9):e0273920. doi: 10.1371/journal.pone.0273920. eCollection 2022. PLoS One. 2022. PMID: 36048783 Free PMC article.
-
Barrier-to-autointegration factor 1 promotes gammaherpesvirus reactivation from latency.Nat Commun. 2023 Feb 6;14(1):434. doi: 10.1038/s41467-023-35898-2. Nat Commun. 2023. PMID: 36746947 Free PMC article.
-
Kaposi's Sarcoma-Associated Herpesvirus Latency-Associated Nuclear Antigen: Replicating and Shielding Viral DNA during Viral Persistence.J Virol. 2017 Jun 26;91(14):e01083-16. doi: 10.1128/JVI.01083-16. Print 2017 Jul 15. J Virol. 2017. PMID: 28446671 Free PMC article. Review.
-
PPM1G restricts innate immune signaling mediated by STING and MAVS and is hijacked by KSHV for immune evasion.Sci Adv. 2020 Nov 20;6(47):eabd0276. doi: 10.1126/sciadv.abd0276. Print 2020 Nov. Sci Adv. 2020. PMID: 33219031 Free PMC article.
References
-
- Kawai T, Akira S. The role of pattern-recognition receptors in innate immunity: update on Toll-like receptors. Nature Immunology. 2010;11:373–384. - PubMed
-
- Janeway CA., Jr. Approaching the asymptote? Evolution and revolution in immunology. Cold Spring Harb.Symp.Quant.Biol. 1989;54(Pt 1):1–13. - PubMed
-
- Poltorak A, He X, Smirnova I, Liu M, van Huffel C, Du X, Birdwell D, Alejos E, Silva M, Galanos C, Freudenberg M, Ricciardi-Castagnoli P, Layton B, Beutler B. Defective LPS signaling in C3H/HeJ and C57BL/10ScCr mice: Mutations in Tlr4 gene. Science. 1999;282:2085–2088. - PubMed
-
- Imai Y, Kuba K, Neely GG, Yaghubian-Malhami R, Perkmann T, van Loo G, Ermolaeva M, Veldhuizen R, Leung YH, Wang H, Liu H, Sun Y, Pasparakis M, Kopf M, Mech C, Bavari S, Peiris JS, Slutsky AS, Akira S, Hultqvist M, Holmdahl R, Nicholls J, Jiang C, Binder CJ, Penninger JM. Identification of oxidative stress and Toll-like receptor 4 signaling as a key pathway of acute lung injury. Cell. 2008;133:235–249. - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

