Phosphorylation variation during the cell cycle scales with structural propensities of proteins
- PMID: 23326221
- PMCID: PMC3542066
- DOI: 10.1371/journal.pcbi.1002842
Phosphorylation variation during the cell cycle scales with structural propensities of proteins
Abstract
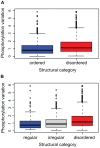
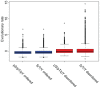
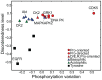
Phosphorylation at specific residues can activate a protein, lead to its localization to particular compartments, be a trigger for protein degradation and fulfill many other biological functions. Protein phosphorylation is increasingly being studied at a large scale and in a quantitative manner that includes a temporal dimension. By contrast, structural properties of identified phosphorylation sites have so far been investigated in a static, non-quantitative way. Here we combine for the first time dynamic properties of the phosphoproteome with protein structural features. At six time points of the cell division cycle we investigate how the variation of the amount of phosphorylation correlates with the protein structure in the vicinity of the modified site. We find two distinct phosphorylation site groups: intrinsically disordered regions tend to contain sites with dynamically varying levels, whereas regions with predominantly regular secondary structures retain more constant phosphorylation levels. The two groups show preferences for different amino acids in their kinase recognition motifs - proline and other disorder-associated residues are enriched in the former group and charged residues in the latter. Furthermore, these preferences scale with the degree of disorderedness, from regular to irregular and to disordered structures. Our results suggest that the structural organization of the region in which a phosphorylation site resides may serve as an additional control mechanism. They also imply that phosphorylation sites are associated with different time scales that serve different functional needs.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures


Similar articles
-
Intrinsic disorder in the Protein Data Bank.J Biomol Struct Dyn. 2007 Feb;24(4):325-42. doi: 10.1080/07391102.2007.10507123. J Biomol Struct Dyn. 2007. PMID: 17206849
-
Characterizing the microenvironment surrounding phosphorylated protein sites.Genomics Proteomics Bioinformatics. 2005 Nov;3(4):213-7. doi: 10.1016/s1672-0229(05)03029-9. Genomics Proteomics Bioinformatics. 2005. PMID: 16689688 Free PMC article.
-
The Nuanced Interplay of Intrinsic Disorder and Other Structural Properties Driving Protein Evolution.Mol Biol Evol. 2016 Sep;33(9):2248-56. doi: 10.1093/molbev/msw092. Epub 2016 May 5. Mol Biol Evol. 2016. PMID: 27189555
-
Modulation of Intrinsically Disordered Protein Function by Post-translational Modifications.J Biol Chem. 2016 Mar 25;291(13):6696-705. doi: 10.1074/jbc.R115.695056. Epub 2016 Feb 5. J Biol Chem. 2016. PMID: 26851279 Free PMC article. Review.
-
Intrinsically unstructured proteins: re-assessing the protein structure-function paradigm.J Mol Biol. 1999 Oct 22;293(2):321-31. doi: 10.1006/jmbi.1999.3110. J Mol Biol. 1999. PMID: 10550212 Review.
Cited by
-
A mechanism of global shape-dependent recognition and phosphorylation of filamin by protein kinase A.J Biol Chem. 2015 Mar 27;290(13):8527-38. doi: 10.1074/jbc.M114.633446. Epub 2015 Feb 9. J Biol Chem. 2015. PMID: 25666618 Free PMC article.
-
Autonomous clocks that regulate organelle biogenesis, cytoskeletal organization, and intracellular dynamics.Elife. 2021 Sep 29;10:e72104. doi: 10.7554/eLife.72104. Elife. 2021. PMID: 34586070 Free PMC article. Review.
-
Digested disorder: Quarterly intrinsic disorder digest (January/February/March, 2013).Intrinsically Disord Proteins. 2013 Apr 1;1(1):e25496. doi: 10.4161/idp.25496. eCollection 2013 Jan-Dec. Intrinsically Disord Proteins. 2013. PMID: 28516015 Free PMC article. Review.
-
Acetylation and phosphorylation control both local and global stability of the chloroplast F1 ATP synthase.Sci Rep. 2017 Mar 9;7:44068. doi: 10.1038/srep44068. Sci Rep. 2017. PMID: 28276484 Free PMC article.
-
Design Principles of Phosphorylation-Dependent Timekeeping in Eukaryotic Circadian Clocks.Cold Spring Harb Perspect Biol. 2018 Aug 1;10(8):a028357. doi: 10.1101/cshperspect.a028357. Cold Spring Harb Perspect Biol. 2018. PMID: 29038116 Free PMC article. Review.
References
-
- Cohen P (2000) The regulation of protein function by multisite phosphorylation–a 25 year update. Trends in biochemical sciences 25: 596–601. - PubMed
-
- Olsen JV, Blagoev B, Gnad F, Macek B, Kumar C, et al. (2006) Global, in vivo, and site-specific phosphorylation dynamics in signaling networks. Cell 127: 635–648. - PubMed
-
- Cox J, Mann M (2008) MaxQuant enables high peptide identification rates, individualized p.p.b.-range mass accuracies and proteome-wide protein quantification. Nature biotechnology 26: 1367–1372. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

