MicroRNA-182-5p targets a network of genes involved in DNA repair
- PMID: 23249749
- PMCID: PMC3543090
- DOI: 10.1261/rna.034926.112
MicroRNA-182-5p targets a network of genes involved in DNA repair
Abstract
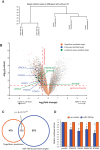
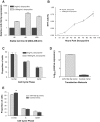
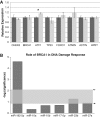
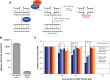
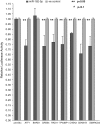
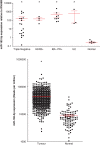
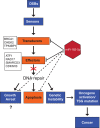
MicroRNAs are noncoding regulators of gene expression, which act by repressing protein translation and/or degrading mRNA. Many have been shown to drive tumorigenesis in cancer, but functional studies to understand their mode of action are typically limited to single-target genes. In this study, we use synthetic biotinylated miRNA to pull down endogenous targets of miR-182-5p. We identified more than 1000 genes as potential targets of miR-182-5p, most of which have a known function in pathways underlying tumor biology. Specifically, functional enrichment analysis identified components of both the DNA damage response pathway and cell cycle to be highly represented in this target cohort. Experimental validation confirmed that miR-182-5p-mediated disruption of the homologous recombination (HR) pathway is a consequence of its ability to target multiple components in that pathway. Although there is a strong enrichment for the cell cycle ontology, we do not see primary proliferative defects as a consequence of miR-182-5p overexpression. We highlight targets that could be responsible for miR-182-5p-mediated disruption of other biological processes attributed in the literature so far. Finally, we show that miR-182-5p is highly expressed in a panel of human breast cancer samples, highlighting its role as a potential oncomir in breast cancer.
Figures

Similar articles
-
miR-151-5p, targeting chromatin remodeler SMARCA5, as a marker for the BRCAness phenotype.Oncotarget. 2016 Dec 6;7(49):80363-80372. doi: 10.18632/oncotarget.10345. Oncotarget. 2016. PMID: 27385001 Free PMC article.
-
miR-7-5p overexpression suppresses cell proliferation and promotes apoptosis through inhibiting the ability of DNA damage repair of PARP-1 and BRCA1 in TK6 cells exposed to hydroquinone.Chem Biol Interact. 2018 Mar 1;283:84-90. doi: 10.1016/j.cbi.2018.01.019. Epub 2018 Feb 5. Chem Biol Interact. 2018. PMID: 29421518
-
Identification of a set of miRNAs differentially expressed in transiently TIA-depleted HeLa cells by genome-wide profiling.BMC Mol Biol. 2013 Feb 6;14:4. doi: 10.1186/1471-2199-14-4. BMC Mol Biol. 2013. PMID: 23387986 Free PMC article.
-
Therapeutic exploitation of tumor cell defects in homologous recombination.Anticancer Agents Med Chem. 2008 May;8(4):448-60. doi: 10.2174/187152008784220267. Anticancer Agents Med Chem. 2008. PMID: 18473729 Review.
-
The role of epigenetic transcriptional regulation in BRCA1-mediated tumor suppression.Transcription. 2013 Jan-Feb;4(1):24-8. doi: 10.4161/trns.22600. Epub 2012 Nov 6. Transcription. 2013. PMID: 23131665 Free PMC article. Review.
Cited by
-
Unraveling the Potential of miRNAs from CSCs as an Emerging Clinical Tool for Breast Cancer Diagnosis and Prognosis.Int J Mol Sci. 2023 Nov 6;24(21):16010. doi: 10.3390/ijms242116010. Int J Mol Sci. 2023. PMID: 37958993 Free PMC article. Review.
-
Identification of specific microRNA-messenger RNA regulation pairs in four subtypes of breast cancer.IET Syst Biol. 2020 Jun;14(3):120-126. doi: 10.1049/iet-syb.2019.0086. IET Syst Biol. 2020. PMID: 32406376 Free PMC article.
-
Cell migration and proliferation are regulated by miR-26a in colorectal cancer via the PTEN-AKT axis.Cancer Cell Int. 2019 Apr 2;19:80. doi: 10.1186/s12935-019-0802-5. eCollection 2019. Cancer Cell Int. 2019. PMID: 30983885 Free PMC article.
-
Transcriptional and post-transcriptional upregulation of p27 mediates growth inhibition of isorhapontigenin (ISO) on human bladder cancer cells.Carcinogenesis. 2018 Mar 8;39(3):482-492. doi: 10.1093/carcin/bgy015. Carcinogenesis. 2018. PMID: 29409027 Free PMC article.
-
Two oncomiRs, miR-182-5p and miR-103a-3p, Involved in Intravenous Leiomyomatosis.Genes (Basel). 2023 Mar 14;14(3):712. doi: 10.3390/genes14030712. Genes (Basel). 2023. PMID: 36980984 Free PMC article.
References
-
- Benjamini Y, Hochberg Y 1995. Controlling the false discovery rate: A practical and powerful approach to multiple testing. J R Stat Soc Series B Stat Methodol 57: 289–300
-
- Bentwich I 2005. Prediction and validation of microRNAs and their targets. FEBS Lett 579: 5904–5910 - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
