LSM proteins provide accurate splicing and decay of selected transcripts to ensure normal Arabidopsis development
- PMID: 23221597
- PMCID: PMC3556967
- DOI: 10.1105/tpc.112.103697
LSM proteins provide accurate splicing and decay of selected transcripts to ensure normal Arabidopsis development
Abstract
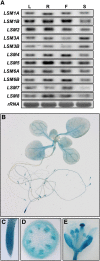
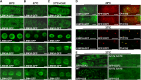
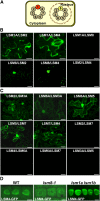
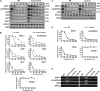
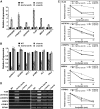
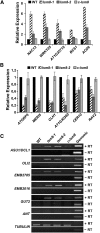
In yeast and animals, SM-like (LSM) proteins typically exist as heptameric complexes and are involved in different aspects of RNA metabolism. Eight LSM proteins, LSM1 to 8, are highly conserved and form two distinct heteroheptameric complexes, LSM1-7 and LSM2-8,that function in mRNA decay and splicing, respectively. A search of the Arabidopsis thaliana genome identifies 11 genes encoding proteins related to the eight conserved LSMs, the genes encoding the putative LSM1, LSM3, and LSM6 proteins being duplicated. Here, we report the molecular and functional characterization of the Arabidopsis LSM gene family. Our results show that the 11 LSM genes are active and encode proteins that are also organized in two different heptameric complexes. The LSM1-7 complex is cytoplasmic and is involved in P-body formation and mRNA decay by promoting decapping. The LSM2-8 complex is nuclear and is required for precursor mRNA splicing through U6 small nuclear RNA stabilization. More importantly, our results also reveal that these complexes are essential for the correct turnover and splicing of selected development-related mRNAs and for the normal development of Arabidopsis. We propose that LSMs play a critical role in Arabidopsis development by ensuring the appropriate development-related gene expression through the regulation of mRNA splicing and decay.
Figures



Similar articles
-
Molecular basis for the distinct cellular functions of the Lsm1-7 and Lsm2-8 complexes.RNA. 2020 Oct;26(10):1400-1413. doi: 10.1261/rna.075879.120. Epub 2020 Jun 9. RNA. 2020. PMID: 32518066 Free PMC article.
-
Arabidopsis thaliana LSM proteins function in mRNA splicing and degradation.Nucleic Acids Res. 2013 Jul;41(12):6232-49. doi: 10.1093/nar/gkt296. Epub 2013 Apr 24. Nucleic Acids Res. 2013. PMID: 23620288 Free PMC article.
-
Emerging Roles of LSM Complexes in Posttranscriptional Regulation of Plant Response to Abiotic Stress.Front Plant Sci. 2019 Feb 19;10:167. doi: 10.3389/fpls.2019.00167. eCollection 2019. Front Plant Sci. 2019. PMID: 30873189 Free PMC article. Review.
-
The sole LSm complex in Cyanidioschyzon merolae associates with pre-mRNA splicing and mRNA degradation factors.RNA. 2017 Jun;23(6):952-967. doi: 10.1261/rna.058487.116. Epub 2017 Mar 21. RNA. 2017. PMID: 28325844 Free PMC article.
-
Pat1 RNA-binding proteins: Multitasking shuttling proteins.Wiley Interdiscip Rev RNA. 2019 Nov;10(6):e1557. doi: 10.1002/wrna.1557. Epub 2019 Jun 24. Wiley Interdiscip Rev RNA. 2019. PMID: 31231973 Review.
Cited by
-
Integrative analysis of the nuclear proteome in Pinus radiata reveals thermopriming coupled to epigenetic regulation.J Exp Bot. 2020 Mar 25;71(6):2040-2057. doi: 10.1093/jxb/erz524. J Exp Bot. 2020. PMID: 31781741 Free PMC article.
-
Role for LSM genes in the regulation of circadian rhythms.Proc Natl Acad Sci U S A. 2014 Oct 21;111(42):15166-71. doi: 10.1073/pnas.1409791111. Epub 2014 Oct 6. Proc Natl Acad Sci U S A. 2014. PMID: 25288739 Free PMC article.
-
Polysomes, Stress Granules, and Processing Bodies: A Dynamic Triumvirate Controlling Cytoplasmic mRNA Fate and Function.Plant Physiol. 2018 Jan;176(1):254-269. doi: 10.1104/pp.17.01468. Epub 2017 Nov 20. Plant Physiol. 2018. PMID: 29158329 Free PMC article. Review.
-
Arabidopsis RNA processing body components LSM1 and DCP5 aid in the evasion of translational repression during Cauliflower mosaic virus infection.Plant Cell. 2022 Jul 30;34(8):3128-3147. doi: 10.1093/plcell/koac132. Plant Cell. 2022. PMID: 35511183 Free PMC article.
-
Composition and function of P bodies in Arabidopsis thaliana.Front Plant Sci. 2014 May 14;5:201. doi: 10.3389/fpls.2014.00201. eCollection 2014. Front Plant Sci. 2014. PMID: 24860588 Free PMC article. Review.
References
-
- Barta A., Kalyna M., Lorković Z.J. (2008). Plant SR proteins and their functions. Curr. Top. Microbiol. Immunol. 326: 83–102 - PubMed
-
- Beelman C.A., Stevens A., Caponigro G., LaGrandeur T.E., Hatfield L., Fortner D.M., Parker R. (1996). An essential component of the decapping enzyme required for normal rates of mRNA turnover. Nature 382: 642–646 - PubMed
-
- Beggs J.D. (2005). Lsm proteins and RNA processing. Biochem. Soc. Trans. 33: 433–438 - PubMed
-
- Belostotsky D.A., Sieburth L.E. (2009). Kill the messenger: mRNA decay and plant development. Curr. Opin. Plant Biol. 12: 96–102 - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

