East African cassava mosaic-like viruses from Africa to Indian ocean islands: molecular diversity, evolutionary history and geographical dissemination of a bipartite begomovirus
- PMID: 23186303
- PMCID: PMC3560262
- DOI: 10.1186/1471-2148-12-228
East African cassava mosaic-like viruses from Africa to Indian ocean islands: molecular diversity, evolutionary history and geographical dissemination of a bipartite begomovirus
Abstract
Background: Cassava (Manihot esculenta) is a major food source for over 200 million sub-Saharan Africans. Unfortunately, its cultivation is severely hampered by cassava mosaic disease (CMD). Caused by a complex of bipartite cassava mosaic geminiviruses (CMG) species (Family: Geminivirideae; Genus: Begomovirus) CMD has been widely described throughout Africa and it is apparent that CMG's are expanding their geographical distribution. Determining where and when CMG movements have occurred could help curtail its spread and reveal the ecological and anthropic factors associated with similar viral invasions. We applied Bayesian phylogeographic inference and recombination analyses to available and newly described CMG sequences to reconstruct a plausible history of CMG diversification and migration between Africa and South West Indian Ocean (SWIO) islands.
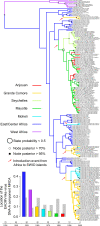
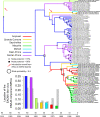
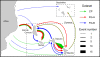
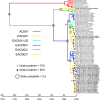
Results: The isolation and analysis of 114 DNA-A and 41 DNA-B sequences demonstrated the presence of three CMG species circulating in the Comoros and Seychelles archipelagos (East African cassava mosaic virus, EACMV; East African cassava mosaic Kenya virus, EACMKV; and East African cassava mosaic Cameroon virus, EACMCV). Phylogeographic analyses suggest that CMG's presence on these SWIO islands is probably the result of at least four independent introduction events from mainland Africa occurring between 1988 and 2009. Amongst the islands of the Comoros archipelago, two major migration pathways were inferred: One from Grande Comore to Mohéli and the second from Mayotte to Anjouan. While only two recombination events characteristic of SWIO islands isolates were identified, numerous re-assortments events were detected between EACMV and EACMKV, which seem to almost freely interchange their genome components.
Conclusions: Rapid and extensive virus spread within the SWIO islands was demonstrated for three CMG complex species. Strong evolutionary or ecological interaction between CMG species may explain both their propensity to exchange components and the absence of recombination with non-CMG begomoviruses. Our results suggest an important role of anthropic factors in CMGs spread as the principal axes of viral migration correspond with major routes of human movement and commercial trade. Finer-scale temporal analyses of CMGs to precisely scale the relative contributions of human and insect transmission to their movement dynamics will require further extensive sampling in the SWIO region.
Figures

Similar articles
-
Divergent evolutionary and epidemiological dynamics of cassava mosaic geminiviruses in Madagascar.BMC Evol Biol. 2016 Sep 6;16(1):182. doi: 10.1186/s12862-016-0749-2. BMC Evol Biol. 2016. PMID: 27600545 Free PMC article.
-
A novel cassava-infecting begomovirus from Madagascar: cassava mosaic Madagascar virus.Arch Virol. 2012 Oct;157(10):2027-30. doi: 10.1007/s00705-012-1399-3. Epub 2012 Jul 10. Arch Virol. 2012. PMID: 22777180
-
Molecular biodiversity of cassava begomoviruses in Tanzania: evolution of cassava geminiviruses in Africa and evidence for East Africa being a center of diversity of cassava geminiviruses.Virol J. 2005 Mar 22;2:21. doi: 10.1186/1743-422X-2-21. Virol J. 2005. PMID: 15784145 Free PMC article.
-
Diversity of dicotyledenous-infecting geminiviruses and their associated DNA molecules in southern Africa, including the South-west Indian ocean islands.Viruses. 2012 Sep;4(9):1753-91. doi: 10.3390/v4091753. Epub 2012 Sep 24. Viruses. 2012. PMID: 23170182 Free PMC article. Review.
-
Cassava mosaic geminiviruses in Africa.Plant Mol Biol. 2004 Nov;56(4):585-99. doi: 10.1007/s11103-004-1651-7. Plant Mol Biol. 2004. PMID: 15630622 Review.
Cited by
-
Mixed Infections of Four Viruses, the Incidence and Phylogenetic Relationships of Sweet Potato Chlorotic Fleck Virus (Betaflexiviridae) Isolates in Wild Species and Sweetpotatoes in Uganda and Evidence of Distinct Isolates in East Africa.PLoS One. 2016 Dec 22;11(12):e0167769. doi: 10.1371/journal.pone.0167769. eCollection 2016. PLoS One. 2016. PMID: 28005969 Free PMC article.
-
Host ecology determines the dispersal patterns of a plant virus.Virus Evol. 2015 Dec 16;1(1):vev016. doi: 10.1093/ve/vev016. eCollection 2015. Virus Evol. 2015. PMID: 27774287 Free PMC article.
-
Synergy between an emerging monopartite begomovirus and a DNA-B component.Sci Rep. 2022 Jan 13;12(1):695. doi: 10.1038/s41598-021-03957-7. Sci Rep. 2022. PMID: 35027584 Free PMC article.
-
Interspecies Recombination Has Driven the Macroevolution of Cassava Mosaic Begomoviruses.J Virol. 2021 Aug 10;95(17):e0054121. doi: 10.1128/JVI.00541-21. Epub 2021 Aug 10. J Virol. 2021. PMID: 34106000 Free PMC article.
-
Epidemiological Analysis of Cassava Mosaic and Brown Streak Diseases, and Bemisia tabaci in the Comoros Islands.Viruses. 2022 Sep 30;14(10):2165. doi: 10.3390/v14102165. Viruses. 2022. PMID: 36298720 Free PMC article.
References
-
- Fauquet C, Fargette D. African cassava mosaic virus: Etiology, Epidemiology, and Control. Plant Dis. 1990;74:404–411. doi: 10.1094/PD-74-0404. - DOI
-
- Jones WO. Manioc in Africa. Stanford: Stanford University Food Research Institute; 1959.
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
LinkOut - more resources
Full Text Sources

