The genetic landscape of mutations in Burkitt lymphoma
- PMID: 23143597
- PMCID: PMC3674561
- DOI: 10.1038/ng.2468
The genetic landscape of mutations in Burkitt lymphoma
Abstract
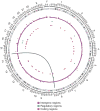
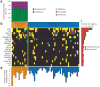
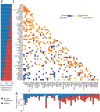
Burkitt lymphoma is characterized by deregulation of MYC, but the contribution of other genetic mutations to the disease is largely unknown. Here, we describe the first completely sequenced genome from a Burkitt lymphoma tumor and germline DNA from the same affected individual. We further sequenced the exomes of 59 Burkitt lymphoma tumors and compared them to sequenced exomes from 94 diffuse large B-cell lymphoma (DLBCL) tumors. We identified 70 genes that were recurrently mutated in Burkitt lymphomas, including ID3, GNA13, RET, PIK3R1 and the SWI/SNF genes ARID1A and SMARCA4. Our data implicate a number of genes in cancer for the first time, including CCT6B, SALL3, FTCD and PC. ID3 mutations occurred in 34% of Burkitt lymphomas and not in DLBCLs. We show experimentally that ID3 mutations promote cell cycle progression and proliferation. Our work thus elucidates commonly occurring gene-coding mutations in Burkitt lymphoma and implicates ID3 as a new tumor suppressor gene.
Figures

Similar articles
-
Recurrent mutation of the ID3 gene in Burkitt lymphoma identified by integrated genome, exome and transcriptome sequencing.Nat Genet. 2012 Dec;44(12):1316-20. doi: 10.1038/ng.2469. Epub 2012 Nov 11. Nat Genet. 2012. PMID: 23143595
-
ID3 mutations are recurrent events in double-hit B-cell lymphomas.Anticancer Res. 2013 Nov;33(11):4771-8. Anticancer Res. 2013. PMID: 24222112
-
The mutational landscape of Burkitt-like lymphoma with 11q aberration is distinct from that of Burkitt lymphoma.Blood. 2019 Feb 28;133(9):962-966. doi: 10.1182/blood-2018-07-864025. Epub 2018 Dec 19. Blood. 2019. PMID: 30567752 Free PMC article.
-
New clues to the molecular pathogenesis of Burkitt lymphoma revealed through next-generation sequencing.Curr Opin Hematol. 2014 Jul;21(4):326-32. doi: 10.1097/MOH.0000000000000059. Curr Opin Hematol. 2014. PMID: 24867287 Review.
-
Oncogenic mechanisms in Burkitt lymphoma.Cold Spring Harb Perspect Med. 2014 Feb 1;4(2):a014282. doi: 10.1101/cshperspect.a014282. Cold Spring Harb Perspect Med. 2014. PMID: 24492847 Free PMC article. Review.
Cited by
-
Clinicopathologic and prognostic relevance of ARID1A protein loss in colorectal cancer.World J Gastroenterol. 2014 Dec 28;20(48):18404-12. doi: 10.3748/wjg.v20.i48.18404. World J Gastroenterol. 2014. PMID: 25561809 Free PMC article.
-
The genetic landscape of high-risk neuroblastoma.Nat Genet. 2013 Mar;45(3):279-84. doi: 10.1038/ng.2529. Epub 2013 Jan 20. Nat Genet. 2013. PMID: 23334666 Free PMC article.
-
The diagnostic gray zone between Burkitt lymphoma and diffuse large B-cell lymphoma is also a gray zone of the mutational spectrum.Leukemia. 2015 Aug;29(8):1789-91. doi: 10.1038/leu.2015.34. Epub 2015 Feb 12. Leukemia. 2015. PMID: 25673238 No abstract available.
-
Distinct Proteomic Profile of Spermatozoa from Men with Seminomatous and Non-Seminomatous Testicular Germ Cell Tumors.Int J Mol Sci. 2020 Jul 8;21(14):4817. doi: 10.3390/ijms21144817. Int J Mol Sci. 2020. PMID: 32650378 Free PMC article.
-
PI3K Isoforms in B Cells.Curr Top Microbiol Immunol. 2022;436:235-254. doi: 10.1007/978-3-031-06566-8_10. Curr Top Microbiol Immunol. 2022. PMID: 36243847
References
-
- Dave SS, et al. Molecular diagnosis of Burkitt's lymphoma. N Engl J Med. 2006;354:2431–2442. - PubMed
-
- Hummel M, et al. A biologic definition of Burkitt's lymphoma from transcriptional and genomic profling. N Engl J Med. 2006;354:2419–2430. - PubMed
-
- Swerdlow SH, et al. WHO Classification of Tumours of Haematopoietic and Lymphoid Tissues. 4th. IARC Press; Lyon, France: 2008.
Publication types
MeSH terms
Substances
Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

