Regulation of the SRC family kinases by Csk
- PMID: 23139636
- PMCID: PMC3492796
- DOI: 10.7150/ijbs.5141
Regulation of the SRC family kinases by Csk
Abstract
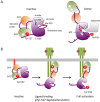
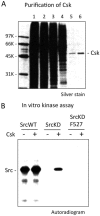
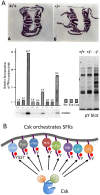
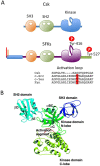
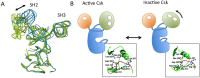
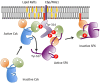
The non-receptor tyrosine kinase Csk serves as an indispensable negative regulator of the Src family tyrosine kinases (SFKs) by specifically phosphorylating the negative regulatory site of SFKs, thereby suppressing their oncogenic potential. Csk is primarily regulated through its SH2 domain, which is required for membrane translocation of Csk via binding to scaffold proteins such as Cbp/PAG1. The binding of scaffolds to the SH2 domain can also upregulate Csk kinase activity. These regulatory features have been elucidated by analyses of Csk structure at the atomic levels. Although Csk itself may not be mutated in human cancers, perturbation of the regulatory system consisting of Csk, Cbp/PAG1, or other scaffolds, and certain tyrosine phosphatases may explain the upregulation of SFKs frequently observed in human cancers. This review focuses on the molecular bases for the function, structure, and regulation of Csk as a unique regulatory tyrosine kinase for SFKs.
Keywords: Csk; Src family; tyrosine kinases.
Conflict of interest statement
Competing Interests: The author has declared that no competing interest exists.
Figures


Similar articles
-
Novel mechanism of regulation of the non-receptor protein tyrosine kinase Csk: insights from NMR mapping studies and site-directed mutagenesis.J Mol Biol. 2001 Nov 16;314(1):129-38. doi: 10.1006/jmbi.2001.5126. J Mol Biol. 2001. PMID: 11724538
-
Detection of a physical and functional interaction between Csk and Lck which involves the SH2 domain of Csk and is mediated by autophosphorylation of Lck on tyrosine 394.J Biol Chem. 1996 Mar 29;271(13):7465-72. doi: 10.1074/jbc.271.13.7465. J Biol Chem. 1996. PMID: 8631775
-
Mechanism of Csk-mediated down-regulation of Src family tyrosine kinases in epidermal growth factor signaling.J Biol Chem. 2004 Feb 13;279(7):5975-83. doi: 10.1074/jbc.M311278200. Epub 2003 Nov 12. J Biol Chem. 2004. PMID: 14613929
-
C-terminal Src kinase (CSK) and CSK-homologous kinase (CHK)--endogenous negative regulators of Src-family protein kinases.Growth Factors. 2005 Sep;23(3):233-44. doi: 10.1080/08977190500178877. Growth Factors. 2005. PMID: 16243715 Review.
-
Structural elements and allosteric mechanisms governing regulation and catalysis of CSK-family kinases and their inhibition of Src-family kinases.Growth Factors. 2010 Oct;28(5):329-50. doi: 10.3109/08977194.2010.484424. Growth Factors. 2010. PMID: 20476842 Review.
Cited by
-
Chemopreventive activity of plant flavonoid isorhamnetin in colorectal cancer is mediated by oncogenic Src and β-catenin.Cancer Res. 2013 Sep 1;73(17):5473-84. doi: 10.1158/0008-5472.CAN-13-0525. Epub 2013 Jul 1. Cancer Res. 2013. PMID: 23824743 Free PMC article.
-
Iron depletion results in Src kinase inhibition with associated cell cycle arrest in neuroblastoma cells.Physiol Rep. 2015 Mar;3(3):e12341. doi: 10.14814/phy2.12341. Physiol Rep. 2015. PMID: 25825542 Free PMC article.
-
Integrin and autocrine IGF2 pathways control fasting insulin secretion in β-cells.J Biol Chem. 2020 Dec 4;295(49):16510-16528. doi: 10.1074/jbc.RA120.012957. Epub 2020 Sep 15. J Biol Chem. 2020. PMID: 32934005 Free PMC article.
-
Chemical signaling in the developing avian retina: Focus on cyclic AMP and AKT-dependent pathways.Front Cell Dev Biol. 2022 Dec 9;10:1058925. doi: 10.3389/fcell.2022.1058925. eCollection 2022. Front Cell Dev Biol. 2022. PMID: 36568967 Free PMC article. Review.
-
Src Kinase Is Biphosphorylated at Y416/Y527 and Activates the CUB-Domain Containing Protein 1/Protein Kinase C δ Pathway in a Subset of Triple-Negative Breast Cancers.Am J Pathol. 2020 Feb;190(2):484-502. doi: 10.1016/j.ajpath.2019.10.017. Epub 2019 Dec 13. Am J Pathol. 2020. PMID: 31843498 Free PMC article.
References
-
- Blume-Jensen P, Hunter T. Oncogenic kinase signalling. Nature. 2001;411:355–65. - PubMed
-
- Brown M, Cooper J. Regulation, substrates and functions of src. Biochim Biophys Acta. 1996;1287:121–49. - PubMed
-
- Thomas S, Brugge J. Cellular Functions regulated by Src Family Kinases. Ann Rev Cell Biol. 1997;13:513–609. - PubMed
-
- Wilde A, Beattie EC, Lem L, Riethof DA, Liu SH, Mobley WC. et al. EGF receptor signaling stimulates SRC kinase phosphorylation of clathrin, influencing clathrin redistribution and EGF uptake. Cell. 1999;96:677–87. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

