MicroRNA and cancer
- PMID: 23102669
- PMCID: PMC5528350
- DOI: 10.1016/j.molonc.2012.09.006
MicroRNA and cancer
Abstract
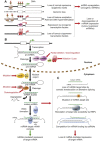
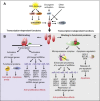
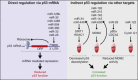
With the advent of next generation sequencing techniques a previously unknown world of non-coding RNA molecules have been discovered. Non-coding RNA transcripts likely outnumber the group of protein coding sequences and hold promise of many new discoveries and mechanistic explanations for essential biological phenomena and pathologies. The best characterized non-coding RNA family consists in humans of about 1400 microRNAs for which abundant evidence have demonstrated fundamental importance in normal development, differentiation, growth control and in human diseases such as cancer. In this review, we summarize the current knowledge and concepts concerning the involvement of microRNAs in cancer, which have emerged from the study of cell culture and animal model systems, including the regulation of key cancer-related pathways, such as cell cycle control and the DNA damage response. Importantly, microRNA molecules are already entering the clinic as diagnostic and prognostic biomarkers for patient stratification and also as therapeutic targets and agents.
Copyright © 2012 Federation of European Biochemical Societies. Published by Elsevier B.V. All rights reserved.
Figures
Similar articles
-
Oncogenesis and Tumor Inhibition by MicroRNAs and its Potential Therapeutic Applications: A Systematic Review.Microrna. 2020;9(3):198-215. doi: 10.2174/2211536608666191104103834. Microrna. 2020. PMID: 31686643
-
MicroRNAs: biogenesis, roles for carcinogenesis and as potential biomarkers for cancer diagnosis and prognosis.Asian Pac J Cancer Prev. 2014;15(18):7489-97. doi: 10.7314/apjcp.2014.15.18.7489. Asian Pac J Cancer Prev. 2014. PMID: 25292018 Review.
-
MicroRNA therapeutics: principles, expectations, and challenges.Chin J Cancer. 2011 Jun;30(6):368-70. doi: 10.5732/cjc.011.10186. Chin J Cancer. 2011. PMID: 21627858 Free PMC article.
-
MicroRNA dysregulation in cancer: diagnostics, monitoring and therapeutics. A comprehensive review.EMBO Mol Med. 2012 Mar;4(3):143-59. doi: 10.1002/emmm.201100209. Epub 2012 Feb 20. EMBO Mol Med. 2012. PMID: 22351564 Free PMC article. Review.
-
Emerging roles of microRNAs as molecular switches in the integrated circuit of the cancer cell.RNA. 2009 Aug;15(8):1443-61. doi: 10.1261/rna.1534709. Epub 2009 Jun 26. RNA. 2009. PMID: 19561119 Free PMC article. Review.
Cited by
-
MicroRNA-93 regulates collagen loss by targeting MMP3 in human nucleus pulposus cells.Cell Prolif. 2015 Jun;48(3):284-92. doi: 10.1111/cpr.12176. Epub 2015 Mar 27. Cell Prolif. 2015. PMID: 25818544 Free PMC article.
-
Paclitaxel Suppresses Hepatocellular Carcinoma Tumorigenesis Through Regulating Circ-BIRC6/miR-877-5p/YWHAZ Axis.Onco Targets Ther. 2020 Sep 22;13:9377-9388. doi: 10.2147/OTT.S261700. eCollection 2020. Onco Targets Ther. 2020. PMID: 33061425 Free PMC article.
-
Cell survival is dicey without Dicer.Mol Cell Oncol. 2014 Jun;1(2):e961825. doi: 10.4161/23723548.2014.961825. Mol Cell Oncol. 2014. PMID: 25558471 Free PMC article.
-
MiR-204 silencing in intraepithelial to invasive cutaneous squamous cell carcinoma progression.Mol Cancer. 2016 Jul 25;15(1):53. doi: 10.1186/s12943-016-0537-z. Mol Cancer. 2016. PMID: 27457246 Free PMC article.
-
Current techniques for visualizing RNA in cells.F1000Res. 2016 Apr 28;5:F1000 Faculty Rev-775. doi: 10.12688/f1000research.8151.1. eCollection 2016. F1000Res. 2016. PMID: 27158473 Free PMC article. Review.
References
-
- Anderson, P. , Kedersha, N. , 2008. Stress granules: the Tao of RNA triage. Trends Biochem. Sci. 33, 141–150. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

