A novel function of RNAs arising from the long terminal repeat of human endogenous retrovirus 9 in cell cycle arrest
- PMID: 23097441
- PMCID: PMC3536363
- DOI: 10.1128/JVI.01648-12
A novel function of RNAs arising from the long terminal repeat of human endogenous retrovirus 9 in cell cycle arrest
Abstract
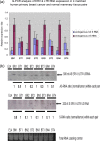
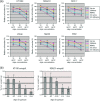
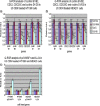
The human genome contains approximately 50 copies of the replication-defective human endogenous retrovirus 9 (ERV-9) and thousands of copies of its solitary long term repeat (sLTR) element. While some sLTRs are located upstream of critical genes and have enhancer activity, other sLTRs are located within introns and may be transcribed as RNAs. We found that intronic RNAs arising from U3 sLTRs of ERV-9 were expressed as both sense (S) and antisense (AS) transcripts in all human cells tested but that expression levels differed in malignant versus nonmalignant cells. In nonmalignant cells, AS was expressed at higher levels than S and at higher levels than in malignant cells; in malignant cells, AS was expressed at amounts equivalent to those of S RNA. Critically, U3 AS RNA was found to physically bind to key transcription factors for cellular proliferation, including NF-Y, p53, and sp1, indicating that such RNA transcripts may function as decoy targets or traps for NF-Y and thus inhibit the growth of human cancer cells. Indeed, short U3 oligodeoxynucleotides (ODNs) based on these RNA sequences ably inhibited proliferation of cancer cell lines driven by cyclins B1/B2, the gene targets of NF-Y.
Figures






Similar articles
-
The long terminal repeat (LTR) of ERV-9 human endogenous retrovirus binds to NF-Y in the assembly of an active LTR enhancer complex NF-Y/MZF1/GATA-2.J Biol Chem. 2005 Oct 21;280(42):35184-94. doi: 10.1074/jbc.M508138200. Epub 2005 Aug 16. J Biol Chem. 2005. PMID: 16105833
-
The solitary long terminal repeats of ERV-9 endogenous retrovirus are conserved during primate evolution and possess enhancer activities in embryonic and hematopoietic cells.J Virol. 2002 Mar;76(5):2410-23. doi: 10.1128/jvi.76.5.2410-2423.2002. J Virol. 2002. PMID: 11836419 Free PMC article.
-
Characterization of molecular clones of porcine endogenous retrovirus-A containing different numbers of U3 repeat boxes in the long terminal repeat region.J Virol Methods. 2012 Apr;181(1):103-8. doi: 10.1016/j.jviromet.2012.01.023. Epub 2012 Feb 11. J Virol Methods. 2012. PMID: 22343070
-
Utility of next-generation RNA-sequencing in identifying chimeric transcription involving human endogenous retroviruses.APMIS. 2016 Jan-Feb;124(1-2):127-39. doi: 10.1111/apm.12477. APMIS. 2016. PMID: 26818267 Review.
-
Endogenous Retroviruses Walk a Fine Line between Priming and Silencing.Viruses. 2020 Jul 23;12(8):792. doi: 10.3390/v12080792. Viruses. 2020. PMID: 32718022 Free PMC article. Review.
Cited by
-
Comprehensive identification of genes driven by ERV9-LTRs reveals TNFRSF10B as a re-activatable mediator of testicular cancer cell death.Cell Death Differ. 2016 Jan;23(1):64-75. doi: 10.1038/cdd.2015.68. Epub 2015 May 29. Cell Death Differ. 2016. PMID: 26024393 Free PMC article.
-
The Sophisticated Transcriptional Response Governed by Transposable Elements in Human Health and Disease.Int J Mol Sci. 2020 Apr 30;21(9):3201. doi: 10.3390/ijms21093201. Int J Mol Sci. 2020. PMID: 32366056 Free PMC article. Review.
-
The intertwining of transposable elements and non-coding RNAs.Int J Mol Sci. 2013 Jun 26;14(7):13307-28. doi: 10.3390/ijms140713307. Int J Mol Sci. 2013. PMID: 23803660 Free PMC article. Review.
-
The animal nuclear factor Y: an enigmatic and important heterotrimeric transcription factor.Am J Cancer Res. 2018 Jul 1;8(7):1106-1125. eCollection 2018. Am J Cancer Res. 2018. PMID: 30094088 Free PMC article. Review.
-
Expressional activation and functional roles of human endogenous retroviruses in cancers.Rev Med Virol. 2019 Mar;29(2):e2025. doi: 10.1002/rmv.2025. Epub 2019 Jan 6. Rev Med Virol. 2019. PMID: 30614117 Free PMC article. Review.
References
-
- Subramanian R, Wildschutte J, Russo C, Coffin J. 2011. Identification, characterization, and comparative genomic distribution of the HERV-K (HML-2) group of human endogenous retroviruses. Retrovirology 8:90 doi:10.1186/1742-4690-8-90 - DOI - PMC - PubMed
-
- Bannert N, Kurth R. 2006. The evolutionary dynamics of human endogenous retroviral families. Annu. Rev. Genomics Hum. Genet. 7:149–173 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

