Dicer partner proteins tune the length of mature miRNAs in flies and mammals
- PMID: 23063653
- PMCID: PMC3609031
- DOI: 10.1016/j.cell.2012.09.027
Dicer partner proteins tune the length of mature miRNAs in flies and mammals
Erratum in
- Cell. 2012 Nov 9;151(4):912
-
Dicer Partner Proteins Tune the Length of Mature miRNAs in Flies and Mammals.Cell. 2012 Nov 9;151(4):912. doi: 10.1016/j.cell.2012.10.029. Epub 2012 Nov 8. Cell. 2012. PMID: 30360291 No abstract available.
Abstract
Drosophila Dicer-1 produces microRNAs (miRNAs) from pre-miRNA, whereas Dicer-2 generates small interfering RNAs (siRNAs) from long dsRNA. Alternative splicing of the loquacious (loqs) mRNA generates three distinct Dicer partner proteins. To understand the function of each, we constructed flies expressing Loqs-PA, Loqs-PB, or Loqs-PD. Loqs-PD promotes both endo- and exo-siRNA production by Dicer-2. Loqs-PA or Loqs-PB is required for viability, but the proteins are not fully redundant: a specific subset of miRNAs requires Loqs-PB. Surprisingly, Loqs-PB tunes where Dicer-1 cleaves pre-miR-307a, generating a longer miRNA isoform with a distinct seed sequence and target specificity. The longer form of miR-307a represses glycerol kinase and taranis mRNA expression. The mammalian Dicer-partner TRBP, a Loqs-PB homolog, similarly tunes where Dicer cleaves pre-miR-132. Thus, Dicer-binding partner proteins change the choice of cleavage site by Dicer, producing miRNAs with target specificities different from those made by Dicer alone or Dicer bound to alternative protein partners.
Copyright © 2012 Elsevier Inc. All rights reserved.
Figures
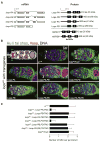

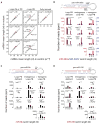

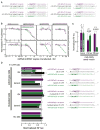
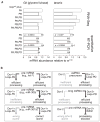
Comment in
-
Dicer partners expand the repertoire of miRNA targets.Genome Biol. 2012 Nov 29;13(11):179. doi: 10.1186/gb-2012-13-11-179. Genome Biol. 2012. PMID: 23194401 Free PMC article.
Similar articles
-
Loquacious-PD removes phosphate inhibition of Dicer-2 processing of hairpin RNAs into siRNAs.Biochem Biophys Res Commun. 2018 Apr 15;498(4):1022-1027. doi: 10.1016/j.bbrc.2018.03.108. Epub 2018 Mar 16. Biochem Biophys Res Commun. 2018. PMID: 29550490 Free PMC article.
-
Dicer partners expand the repertoire of miRNA targets.Genome Biol. 2012 Nov 29;13(11):179. doi: 10.1186/gb-2012-13-11-179. Genome Biol. 2012. PMID: 23194401 Free PMC article.
-
Dicer partner protein tunes the length of miRNAs using base-mismatch in the pre-miRNA stem.Nucleic Acids Res. 2018 Apr 20;46(7):3726-3741. doi: 10.1093/nar/gky043. Nucleic Acids Res. 2018. PMID: 29373753 Free PMC article.
-
MicroRNAs: Loquacious speaks out.Curr Biol. 2005 Aug 9;15(15):R603-5. doi: 10.1016/j.cub.2005.07.044. Curr Biol. 2005. PMID: 16085484 Review.
-
The RNAi pathway initiated by Dicer-2 in Drosophila.Cold Spring Harb Symp Quant Biol. 2006;71:39-44. doi: 10.1101/sqb.2006.71.008. Cold Spring Harb Symp Quant Biol. 2006. PMID: 17381278 Review.
Cited by
-
The Effect of Dicer Knockout on RNA Interference Using Various Dicer Substrate Small Interfering RNA (DsiRNA) Structures.Genes (Basel). 2022 Feb 27;13(3):436. doi: 10.3390/genes13030436. Genes (Basel). 2022. PMID: 35327991 Free PMC article.
-
Double-Stranded RNA Sensors and Modulators in Innate Immunity.Annu Rev Immunol. 2019 Apr 26;37:349-375. doi: 10.1146/annurev-immunol-042718-041356. Epub 2019 Jan 23. Annu Rev Immunol. 2019. PMID: 30673536 Free PMC article. Review.
-
TRBP ensures efficient Dicer processing of precursor microRNA in RNA-crowded environments.Nat Commun. 2016 Dec 9;7:13694. doi: 10.1038/ncomms13694. Nat Commun. 2016. PMID: 27934859 Free PMC article.
-
Nucleotide sequence of miRNA precursor contributes to cleavage site selection by Dicer.Nucleic Acids Res. 2015 Dec 15;43(22):10939-51. doi: 10.1093/nar/gkv968. Epub 2015 Sep 30. Nucleic Acids Res. 2015. PMID: 26424848 Free PMC article.
-
Towards Antiviral shRNAs Based on the AgoshRNA Design.PLoS One. 2015 Jun 18;10(6):e0128618. doi: 10.1371/journal.pone.0128618. eCollection 2015. PLoS One. 2015. PMID: 26087209 Free PMC article.
References
Publication types
MeSH terms
Substances
Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

