(+)RNA viruses rewire cellular pathways to build replication organelles
- PMID: 23036609
- PMCID: PMC7102821
- DOI: 10.1016/j.coviro.2012.09.006
(+)RNA viruses rewire cellular pathways to build replication organelles
Abstract
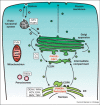
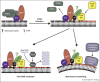
Positive-strand RNA [(+)RNA] viruses show a significant degree of conservation of their mechanisms of replication. The universal requirement of (+)RNA viruses for cellular membranes for genome replication, and the formation of membranous replication organelles with similar architecture, suggest that they target essential control mechanisms of membrane metabolism conserved among eukaryotes. Recently, significant progress has been made in understanding the role of key host factors and pathways that are hijacked for the development of replication organelles. In addition, electron tomography studies have shed new light on their ultrastructure. Collectively, these studies reveal an unexpected complexity of the spatial organization of the replication membranes and suggest that (+)RNA viruses actively change cellular membrane composition to build their replication organelles.
Copyright © 2012 Elsevier B.V. All rights reserved.
Figures

Similar articles
-
Ultrastructural Features of Membranous Replication Organelles Induced by Positive-Stranded RNA Viruses.Cells. 2021 Sep 13;10(9):2407. doi: 10.3390/cells10092407. Cells. 2021. PMID: 34572055 Free PMC article. Review.
-
Building Viral Replication Organelles: Close Encounters of the Membrane Types.PLoS Pathog. 2016 Oct 27;12(10):e1005912. doi: 10.1371/journal.ppat.1005912. eCollection 2016 Oct. PLoS Pathog. 2016. PMID: 27788266 Free PMC article. Review. No abstract available.
-
Composition of plant virus RNA replicase complexes.Curr Opin Virol. 2012 Dec;2(6):669-75. doi: 10.1016/j.coviro.2012.09.014. Epub 2012 Oct 18. Curr Opin Virol. 2012. PMID: 23083891 Review.
-
Host endomembrane recruitment for plant RNA virus replication.Curr Opin Virol. 2012 Dec;2(6):683-90. doi: 10.1016/j.coviro.2012.10.003. Epub 2012 Nov 2. Curr Opin Virol. 2012. PMID: 23123078 Free PMC article. Review.
-
Interaction of the innate immune system with positive-strand RNA virus replication organelles.Cytokine Growth Factor Rev. 2017 Oct;37:17-27. doi: 10.1016/j.cytogfr.2017.05.007. Epub 2017 Jun 27. Cytokine Growth Factor Rev. 2017. PMID: 28709747 Free PMC article.
Cited by
-
Enterovirus Replication Organelles and Inhibitors of Their Formation.Front Microbiol. 2020 Aug 20;11:1817. doi: 10.3389/fmicb.2020.01817. eCollection 2020. Front Microbiol. 2020. PMID: 32973693 Free PMC article. Review.
-
Cellular ESCRT components are recruited to regulate the endocytic trafficking and RNA replication compartment assembly during classical swine fever virus infection.PLoS Pathog. 2022 Feb 4;18(2):e1010294. doi: 10.1371/journal.ppat.1010294. eCollection 2022 Feb. PLoS Pathog. 2022. PMID: 35120190 Free PMC article.
-
Phospholipid synthesis fueled by lipid droplets drives the structural development of poliovirus replication organelles.PLoS Pathog. 2018 Aug 27;14(8):e1007280. doi: 10.1371/journal.ppat.1007280. eCollection 2018 Aug. PLoS Pathog. 2018. PMID: 30148882 Free PMC article.
-
Membrane binding and rearrangement by chikungunya virus capping enzyme nsP1.Virology. 2020 May;544:31-41. doi: 10.1016/j.virol.2020.02.006. Epub 2020 Feb 24. Virology. 2020. PMID: 32174512 Free PMC article.
-
Seneca Valley virus 3Cpro antagonizes host innate immune responses and programmed cell death.Front Microbiol. 2023 Oct 6;14:1235620. doi: 10.3389/fmicb.2023.1235620. eCollection 2023. Front Microbiol. 2023. PMID: 37869659 Free PMC article. Review.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

