Epithelial-to-mesenchymal transition (EMT) confers primary resistance to trastuzumab (Herceptin)
- PMID: 22992620
- PMCID: PMC3507497
- DOI: 10.4161/cc.22225
Epithelial-to-mesenchymal transition (EMT) confers primary resistance to trastuzumab (Herceptin)
Abstract
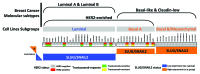
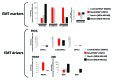
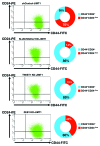
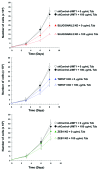
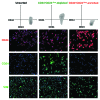
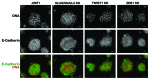
The rate of inherent resistance to single-agent trastuzumab in HER2-overexpressing metastatic breast carcinomas is impressive at above 70%. Unfortunately, little is known regarding the distinctive genetic signatures that could predict trastuzumab refractoriness ab initio. The epithelial-to-mesenchymal transition (EMT) molecular features, HER2 expression status and primary responses to trastuzumab were explored in the public Lawrence Berkeley Laboratory (LBL) Breast Cancer Collection. Lentivirus-delivered small hairpin RNAs were employed to reduce specifically and stably the expression of EMT transcription factors in trastuzumab-refractory basal/HER2+ cells. Cell proliferation assays and pre-clinical nude mice xenograft-based studies were performed to assess the contribution of specific EMT transcription factors to inherent trastuzumab resistance. Primary sensitivity to trastuzumab was restricted to the SLUG/SNAIL2-negative subset of luminal/HER2+ cell lines, whereas all of the SLUG/SNAIL2-positive basal/HER2+ cell lines exhibited an inherent resistance to trastuzumab. The specific knockdown of SLUG/SNAIL2 suppressed the stem-related CD44+CD24(-/low) mesenchymal immunophenotype by transcriptionally upregulating the luminal epithelial marker CD24 in basal/HER2+ cells. Basal/HER2+ cells gained sensitivity to the growth-inhibitory effects of trastuzumab following SLUG/SNAIL2 gene depletion, which induced the expression of the mesenchymal-to-epithelial transition (MET) genes involved in promoting an epithelial phenotype. The isolation of CD44+CD24(-/low) mesenchymal cells by magnetic-activated cell sorting (MACS) confirmed their intrinsic unresponsiveness to trastuzumab. A reduction in tumor growth and dramatic gain in sensitivity to trastuzumab in vivo were confirmed when the SLUG/SNAIL2 knockdown basal/HER2+ cells were injected into nude mice. HER2 overexpression in a basal, rather than in a luminal molecular background, results in a basal/HER2+ breast cancer subtype that is intrinsically resistant to trastuzumab. EMT transcription factors might induce an enhanced phenotypic plasticity that would allow basal/HER2+ breast cancer cells to "enter" into and "exit" dynamically from trastuzumab-responsive stem cell-like states. The systematic determination of SLUG/SNAIL2 as a stem/CD44+CD24(-/low) cell-associated protein may improve the therapeutic management of HER2+ breast carcinomas.
Figures


Similar articles
-
Metformin-induced preferential killing of breast cancer initiating CD44+CD24-/low cells is sufficient to overcome primary resistance to trastuzumab in HER2+ human breast cancer xenografts.Oncotarget. 2012 Apr;3(4):395-8. doi: 10.18632/oncotarget.488. Oncotarget. 2012. PMID: 22565037 Free PMC article.
-
Basal/HER2 breast carcinomas: integrating molecular taxonomy with cancer stem cell dynamics to predict primary resistance to trastuzumab (Herceptin).Cell Cycle. 2013 Jan 15;12(2):225-45. doi: 10.4161/cc.23274. Epub 2012 Jan 15. Cell Cycle. 2013. PMID: 23255137 Free PMC article.
-
Spontaneous epithelial-mesenchymal transition and resistance to HER-2-targeted therapies in HER-2-positive luminal breast cancer.PLoS One. 2013 Aug 26;8(8):e71987. doi: 10.1371/journal.pone.0071987. eCollection 2013. PLoS One. 2013. PMID: 23991019 Free PMC article.
-
Epithelial mesenchymal transition traits in human breast cancer cell lines parallel the CD44(hi/)CD24 (lo/-) stem cell phenotype in human breast cancer.J Mammary Gland Biol Neoplasia. 2010 Jun;15(2):235-52. doi: 10.1007/s10911-010-9175-z. Epub 2010 Jun 4. J Mammary Gland Biol Neoplasia. 2010. PMID: 20521089 Review.
-
Current status of anti-human epidermal growth factor receptor 2 therapies: predicting and overcoming herceptin resistance.Clin Breast Cancer. 2013 Aug;13(4):223-32. doi: 10.1016/j.clbc.2013.04.001. Clin Breast Cancer. 2013. PMID: 23829888 Free PMC article. Review.
Cited by
-
The Autophagic Route of E-Cadherin and Cell Adhesion Molecules in Cancer Progression.Cancers (Basel). 2021 Dec 16;13(24):6328. doi: 10.3390/cancers13246328. Cancers (Basel). 2021. PMID: 34944948 Free PMC article. Review.
-
Is cancer an intelligent species?Cancer Metastasis Rev. 2023 Dec;42(4):1201-1218. doi: 10.1007/s10555-023-10123-0. Epub 2023 Aug 4. Cancer Metastasis Rev. 2023. PMID: 37540301 Free PMC article. Review.
-
Methodology to analyze gene expression patterns of early mammary development in pig models.Mol Biol Rep. 2020 Apr;47(4):3241-3248. doi: 10.1007/s11033-020-05362-1. Epub 2020 Mar 26. Mol Biol Rep. 2020. PMID: 32219771
-
The role of epithelial-mesenchymal transition-regulating transcription factors in anti-cancer drug resistance.Arch Pharm Res. 2021 Mar;44(3):281-292. doi: 10.1007/s12272-021-01321-x. Epub 2021 Mar 25. Arch Pharm Res. 2021. PMID: 33768509 Free PMC article. Review.
-
Preclinical Characteristics of the Irreversible Pan-HER Kinase Inhibitor Neratinib Compared with Lapatinib: Implications for the Treatment of HER2-Positive and HER2-Mutated Breast Cancer.Cancers (Basel). 2019 May 28;11(6):737. doi: 10.3390/cancers11060737. Cancers (Basel). 2019. PMID: 31141894 Free PMC article. Review.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous
