Efficient and sequence-independent replication of DNA containing a third base pair establishes a functional six-letter genetic alphabet
- PMID: 22773812
- PMCID: PMC3409741
- DOI: 10.1073/pnas.1205176109
Efficient and sequence-independent replication of DNA containing a third base pair establishes a functional six-letter genetic alphabet
Abstract
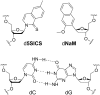
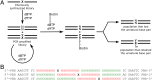
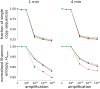
The natural four-letter genetic alphabet, comprised of just two base pairs (dA-dT and dG-dC), is conserved throughout all life, and its expansion by the development of a third, unnatural base pair has emerged as a central goal of chemical and synthetic biology. We recently developed a class of candidate unnatural base pairs, exemplified by the pair formed between d5SICS and dNaM. Here, we examine the PCR amplification of DNA containing one or more d5SICS-dNaM pairs in a wide variety of sequence contexts. Under standard conditions, we show that this DNA may be amplified with high efficiency and greater than 99.9% fidelity. To more rigorously explore potential sequence effects, we used deep sequencing to characterize a library of templates containing the unnatural base pair as a function of amplification. We found that the unnatural base pair is efficiently replicated with high fidelity in virtually all sequence contexts. The results show that, for PCR and PCR-based applications, d5SICS-dNaM is functionally equivalent to a natural base pair, and when combined with dA-dT and dG-dC, it provides a fully functional six-letter genetic alphabet.
Conflict of interest statement
The authors declare no conflict of interest.
Figures

Similar articles
-
Site-Specific Labeling of DNA via PCR with an Expanded Genetic Alphabet.Methods Mol Biol. 2019;1973:193-212. doi: 10.1007/978-1-4939-9216-4_13. Methods Mol Biol. 2019. PMID: 31016704
-
Can a Six-Letter Alphabet Increase the Likelihood of Photochemical Assault to the Genetic Code?Chemistry. 2016 Nov 7;22(46):16648-16656. doi: 10.1002/chem.201602160. Epub 2016 Oct 10. Chemistry. 2016. PMID: 27723147
-
Transcription of an expanded genetic alphabet.J Am Chem Soc. 2009 Apr 15;131(14):5046-7. doi: 10.1021/ja9006996. J Am Chem Soc. 2009. PMID: 19351201 Free PMC article.
-
The expanded genetic alphabet.Angew Chem Int Ed Engl. 2015 Oct 5;54(41):11930-44. doi: 10.1002/anie.201502890. Epub 2015 Aug 25. Angew Chem Int Ed Engl. 2015. PMID: 26304162 Free PMC article. Review.
-
Creation of unnatural base pairs for genetic alphabet expansion toward synthetic xenobiology.Curr Opin Chem Biol. 2018 Oct;46:108-114. doi: 10.1016/j.cbpa.2018.07.017. Epub 2018 Jul 27. Curr Opin Chem Biol. 2018. PMID: 30059833 Review.
Cited by
-
Preparing synthetic biology for the world.Front Microbiol. 2013 Jan 25;4:5. doi: 10.3389/fmicb.2013.00005. eCollection 2013. Front Microbiol. 2013. PMID: 23355834 Free PMC article.
-
Ribonucleosides for an artificially expanded genetic information system.J Org Chem. 2014 Apr 4;79(7):3194-9. doi: 10.1021/jo402665d. Epub 2014 Mar 18. J Org Chem. 2014. PMID: 24597611 Free PMC article.
-
Sequencing for oxidative DNA damage at single-nucleotide resolution with click-code-seq v2.0.Chem Commun (Camb). 2023 Jul 18;59(58):8997-9000. doi: 10.1039/d3cc02699j. Chem Commun (Camb). 2023. PMID: 37401666 Free PMC article.
-
Site-specifically arraying small molecules or proteins on DNA using an expanded genetic alphabet.Chemistry. 2013 Oct 11;19(42):14205-14209. doi: 10.1002/chem.201302496. Epub 2013 Sep 11. Chemistry. 2013. PMID: 24026962 Free PMC article.
-
Overlapping genes in natural and engineered genomes.Nat Rev Genet. 2022 Mar;23(3):154-168. doi: 10.1038/s41576-021-00417-w. Epub 2021 Oct 5. Nat Rev Genet. 2022. PMID: 34611352 Free PMC article. Review.
References
-
- Kawai R, et al. Site-specific fluorescent labeling of RNA molecules by specific transcription using unnatural base pairs. J Am Chem Soc. 2005;127:17286–17295. - PubMed
-
- Kimoto M, et al. A new unnatural base pair system between fluorophore and quencher base analogues for nucleic acid-based imaging technology. J Am Chem Soc. 2010;132:15418–15426. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

