Evolutionary dynamics of genome segmentation in multipartite viruses
- PMID: 22764164
- PMCID: PMC3415918
- DOI: 10.1098/rspb.2012.1086
Evolutionary dynamics of genome segmentation in multipartite viruses
Abstract
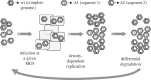
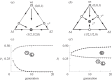
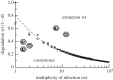
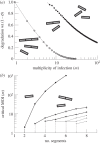
Multipartite viruses are formed by a variable number of genomic fragments packed in independent viral capsids. This fact poses stringent conditions on their transmission mode, demanding, in particular, a high multiplicity of infection (MOI) for successful propagation. The actual advantages of the multipartite viral strategy are as yet unclear. The origin of multipartite viruses represents an evolutionary puzzle. While classical theories suggested that a faster replication rate or higher replication fidelity would favour shorter segments, recent experimental results seem to point to an increased stability of virions with incomplete genomes as a factor able to compensate for the disadvantage of mandatory complementation. Using as main parameters differential stability as a function of genome length and MOI, we calculate the conditions under which a set of complementary segments of a viral genome would outcompete the non-segmented variant. Further, we examine the likeliness that multipartite viral forms could be the evolutionary outcome of the competition among the defective genomes of different lengths that spontaneously arise under replication of a complete, wild-type genome. We conclude that only multipartite viruses with a small number of segments could be produced in our scenario, and discuss alternative hypotheses for the origin of multipartite viruses with more than four segments.
Figures
Similar articles
-
Small Bottleneck Size in a Highly Multipartite Virus during a Complete Infection Cycle.J Virol. 2018 Jun 29;92(14):e00139-18. doi: 10.1128/JVI.00139-18. Print 2018 Jul 15. J Virol. 2018. PMID: 29720515 Free PMC article.
-
Theoretical approaches to disclosing the emergence and adaptive advantages of multipartite viruses.Curr Opin Virol. 2018 Dec;33:89-95. doi: 10.1016/j.coviro.2018.07.018. Epub 2018 Aug 16. Curr Opin Virol. 2018. PMID: 30121469 Review.
-
Cheating leads to the evolution of multipartite viruses.PLoS Biol. 2023 Apr 24;21(4):e3002092. doi: 10.1371/journal.pbio.3002092. eCollection 2023 Apr. PLoS Biol. 2023. PMID: 37093882 Free PMC article.
-
The Curious Strategy of Multipartite Viruses.Annu Rev Virol. 2020 Sep 29;7(1):203-218. doi: 10.1146/annurev-virology-010220-063346. Annu Rev Virol. 2020. PMID: 32991271 Review.
-
Viral genome segmentation can result from a trade-off between genetic content and particle stability.PLoS Genet. 2011 Mar;7(3):e1001344. doi: 10.1371/journal.pgen.1001344. Epub 2011 Mar 17. PLoS Genet. 2011. PMID: 21437265 Free PMC article.
Cited by
-
A multicellular way of life for a multipartite virus.Elife. 2019 Mar 12;8:e43599. doi: 10.7554/eLife.43599. Elife. 2019. PMID: 30857590 Free PMC article.
-
Small Bottleneck Size in a Highly Multipartite Virus during a Complete Infection Cycle.J Virol. 2018 Jun 29;92(14):e00139-18. doi: 10.1128/JVI.00139-18. Print 2018 Jul 15. J Virol. 2018. PMID: 29720515 Free PMC article.
-
Analysis of intra-host genetic diversity of Prunus necrotic ringspot virus (PNRSV) using amplicon next generation sequencing.PLoS One. 2017 Jun 20;12(6):e0179284. doi: 10.1371/journal.pone.0179284. eCollection 2017. PLoS One. 2017. PMID: 28632759 Free PMC article.
-
Multipartite viruses: adaptive trick or evolutionary treat?NPJ Syst Biol Appl. 2017 Nov 9;3:34. doi: 10.1038/s41540-017-0035-y. eCollection 2017. NPJ Syst Biol Appl. 2017. PMID: 29263796 Free PMC article. Review.
-
The Strange Lifestyle of Multipartite Viruses.PLoS Pathog. 2016 Nov 3;12(11):e1005819. doi: 10.1371/journal.ppat.1005819. eCollection 2016 Nov. PLoS Pathog. 2016. PMID: 27812219 Free PMC article. Review.
References
-
- Eigen M. 1993. Viral quasispecies. Scient. Am. 296, 32–39 - PubMed
-
- Roosinck M. J. 1997. Mechanisms of plant virus evolution. Annu. Rev. Phytopathol. 35, 191–209 (doi:10.1146/annurev.phyto.35.1.191) - DOI - PubMed
-
- Manrubia S. C., Lázaro E. 2006. Viral evolution. Phys. Life Rev. 3, 65–92 (doi:10.1016/j.plrev.2005.11.002) - DOI
-
- Szathmary E. 1992. Natural selection and dynamical coexistence of defective and complementing virus segments. J. Theoret. Biol. 157, 383–406 (doi:10.1016/S0022-5193(05)80617-4) - DOI - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources

