MicroRNA profiling: approaches and considerations
- PMID: 22510765
- PMCID: PMC4517822
- DOI: 10.1038/nrg3198
MicroRNA profiling: approaches and considerations
Abstract
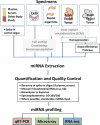
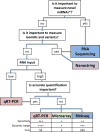
MicroRNAs (miRNAs) are small RNAs that post-transcriptionally regulate the expression of thousands of genes in a broad range of organisms in both normal physiological contexts and in disease contexts. miRNA expression profiling is gaining popularity because miRNAs, as key regulators in gene expression networks, can influence many biological processes and also show promise as biomarkers for disease. Technological advances have spawned a multitude of platforms for miRNA profiling, and an understanding of the strengths and pitfalls of different approaches can aid in their effective use. Here, we review the major considerations for carrying out and interpreting results of miRNA-profiling studies.
Conflict of interest statement
Figures


Similar articles
-
Identification of Taxus microRNAs and their targets with high-throughput sequencing and degradome analysis.Physiol Plant. 2012 Dec;146(4):388-403. doi: 10.1111/j.1399-3054.2012.01668.x. Epub 2012 Jul 25. Physiol Plant. 2012. PMID: 22708792
-
Next-generation sequencing identifies the natural killer cell microRNA transcriptome.Genome Res. 2010 Nov;20(11):1590-604. doi: 10.1101/gr.107995.110. Epub 2010 Oct 8. Genome Res. 2010. PMID: 20935160 Free PMC article.
-
Identification and characterization of microRNAs from peanut (Arachis hypogaea L.) by high-throughput sequencing.PLoS One. 2011;6(11):e27530. doi: 10.1371/journal.pone.0027530. Epub 2011 Nov 16. PLoS One. 2011. PMID: 22110666 Free PMC article.
-
Potential pitfalls in microRNA profiling.Wiley Interdiscip Rev RNA. 2012 Sep-Oct;3(5):601-16. doi: 10.1002/wrna.1120. Epub 2012 May 7. Wiley Interdiscip Rev RNA. 2012. PMID: 22566380 Free PMC article. Review.
-
Methodologies for high-throughput expression profiling of microRNAs.Methods Mol Biol. 2006;342:139-57. doi: 10.1385/1-59745-123-1:139. Methods Mol Biol. 2006. PMID: 16957373 Review.
Cited by
-
Comparison of Methods for miRNA Extraction from Plasma and Quantitative Recovery of RNA from Cerebrospinal Fluid.Front Genet. 2013 May 16;4:83. doi: 10.3389/fgene.2013.00083. eCollection 2013. Front Genet. 2013. PMID: 23720669 Free PMC article.
-
Regulation of B-cell development and function by microRNAs.Immunol Rev. 2013 May;253(1):25-39. doi: 10.1111/imr.12046. Immunol Rev. 2013. PMID: 23550636 Free PMC article. Review.
-
Non-coding RNAs in cancer: platforms and strategies for investigating the genomic "dark matter".J Exp Clin Cancer Res. 2020 Jun 20;39(1):117. doi: 10.1186/s13046-020-01622-x. J Exp Clin Cancer Res. 2020. PMID: 32563270 Free PMC article. Review.
-
miR-31-5p from placental and peripheral blood exosomes is a potential biomarker to diagnose preeclampsia.Hereditas. 2022 Sep 19;159(1):35. doi: 10.1186/s41065-022-00250-z. Hereditas. 2022. PMID: 36123601 Free PMC article.
-
Development of an undergraduate bioinformatics degree program at a liberal arts college.Yale J Biol Med. 2012 Sep;85(3):309-21. Epub 2012 Sep 25. Yale J Biol Med. 2012. PMID: 23012579 Free PMC article.
References
-
- Lee RC, Feinbaum RL, Ambros V. The C. elegans heterochronic gene lin-4 encodes small RNAs with antisense complementarity to lin-14. Cell. 1993;75:843–54. - PubMed
-
- Wightman B, Ha I, Ruvkun G. Posttranscriptional regulation of the heterochronic gene lin-14 by lin-4 mediates temporal pattern formation in C. elegans. Cell. 1993;75:855–62. - PubMed
-
- Reinhart BJ, et al. The 21-nucleotide let-7 RNA regulates developmental timing in Caenorhabditis elegans. Nature. 2000;403:901–6. - PubMed
-
- Kim VN, Han J, Siomi MC. Biogenesis of small RNAs in animals. Nature reviews. Molecular cell biology. 2009;10:126–39. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

