2'-O methylation of internal adenosine by flavivirus NS5 methyltransferase
- PMID: 22496660
- PMCID: PMC3320599
- DOI: 10.1371/journal.ppat.1002642
2'-O methylation of internal adenosine by flavivirus NS5 methyltransferase
Abstract
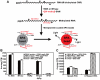
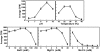
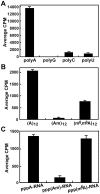
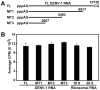
RNA modification plays an important role in modulating host-pathogen interaction. Flavivirus NS5 protein encodes N-7 and 2'-O methyltransferase activities that are required for the formation of 5' type I cap (m(7)GpppAm) of viral RNA genome. Here we reported, for the first time, that flavivirus NS5 has a novel internal RNA methylation activity. Recombinant NS5 proteins of West Nile virus and Dengue virus (serotype 4; DENV-4) specifically methylates polyA, but not polyG, polyC, or polyU, indicating that the methylation occurs at adenosine residue. RNAs with internal adenosines substituted with 2'-O-methyladenosines are not active substrates for internal methylation, whereas RNAs with adenosines substituted with N⁶-methyladenosines can be efficiently methylated, suggesting that the internal methylation occurs at the 2'-OH position of adenosine. Mass spectroscopic analysis further demonstrated that the internal methylation product is 2'-O-methyladenosine. Importantly, genomic RNA purified from DENV virion contains 2'-O-methyladenosine. The 2'-O methylation of internal adenosine does not require specific RNA sequence since recombinant methyltransferase of DENV-4 can efficiently methylate RNAs spanning different regions of viral genome, host ribosomal RNAs, and polyA. Structure-based mutagenesis results indicate that K61-D146-K181-E217 tetrad of DENV-4 methyltransferase forms the active site of internal methylation activity; in addition, distinct residues within the methyl donor (S-adenosyl-L-methionine) pocket, GTP pocket, and RNA-binding site are critical for the internal methylation activity. Functional analysis using flavivirus replicon and genome-length RNAs showed that internal methylation attenuated viral RNA translation and replication. Polymerase assay revealed that internal 2'-O-methyladenosine reduces the efficiency of RNA elongation. Collectively, our results demonstrate that flavivirus NS5 performs 2'-O methylation of internal adenosine of viral RNA in vivo and host ribosomal RNAs in vitro.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures


Similar articles
-
Molecular basis for specific viral RNA recognition and 2'-O-ribose methylation by the dengue virus nonstructural protein 5 (NS5).Proc Natl Acad Sci U S A. 2015 Dec 1;112(48):14834-9. doi: 10.1073/pnas.1514978112. Epub 2015 Nov 17. Proc Natl Acad Sci U S A. 2015. PMID: 26578813 Free PMC article.
-
Structure and function of flavivirus NS5 methyltransferase.J Virol. 2007 Apr;81(8):3891-903. doi: 10.1128/JVI.02704-06. Epub 2007 Jan 31. J Virol. 2007. PMID: 17267492 Free PMC article.
-
Mutagenesis of the dengue virus type 2 NS5 methyltransferase domain.J Biol Chem. 2008 Jul 11;283(28):19410-21. doi: 10.1074/jbc.M800613200. Epub 2008 May 9. J Biol Chem. 2008. PMID: 18469001
-
The Transactions of NS3 and NS5 in Flaviviral RNA Replication.Adv Exp Med Biol. 2018;1062:147-163. doi: 10.1007/978-981-10-8727-1_11. Adv Exp Med Biol. 2018. PMID: 29845531 Review.
-
Flavivirus methyltransferase: a novel antiviral target.Antiviral Res. 2008 Oct;80(1):1-10. doi: 10.1016/j.antiviral.2008.05.003. Epub 2008 Jun 5. Antiviral Res. 2008. PMID: 18571739 Free PMC article. Review.
Cited by
-
Physiologic RNA targets and refined sequence specificity of coronavirus EndoU.RNA. 2020 Dec;26(12):1976-1999. doi: 10.1261/rna.076604.120. Epub 2020 Sep 28. RNA. 2020. PMID: 32989044 Free PMC article.
-
Dynamics of Human and Viral RNA Methylation during Zika Virus Infection.Cell Host Microbe. 2016 Nov 9;20(5):666-673. doi: 10.1016/j.chom.2016.10.002. Epub 2016 Oct 20. Cell Host Microbe. 2016. PMID: 27773536 Free PMC article.
-
Evasion of early innate immune response by 2'-O-methylation of dengue genomic RNA.Virology. 2016 Dec;499:259-266. doi: 10.1016/j.virol.2016.09.022. Epub 2016 Oct 4. Virology. 2016. PMID: 27716465 Free PMC article.
-
Molecular basis for specific viral RNA recognition and 2'-O-ribose methylation by the dengue virus nonstructural protein 5 (NS5).Proc Natl Acad Sci U S A. 2015 Dec 1;112(48):14834-9. doi: 10.1073/pnas.1514978112. Epub 2015 Nov 17. Proc Natl Acad Sci U S A. 2015. PMID: 26578813 Free PMC article.
-
Coronavirus takeover of host cell translation and intracellular antiviral response: a molecular perspective.EMBO J. 2024 Jan;43(2):151-167. doi: 10.1038/s44318-023-00019-8. Epub 2024 Jan 10. EMBO J. 2024. PMID: 38200146 Free PMC article. Review.
References
-
- Gubler D, Kuno G, Markoff L. Flaviviruses. In: Knipe DM, Howley PM, editors. Fields virology, 5th edition. Philadelphia, PA: Lippincott William & Wilkins; 2007. pp. 1153–1253.
-
- Lindenbach BD, Thiel H-J, Rice CM. Flaviviridae: The Virus and Their Replication. In: Knipe DM, Howleym PM, editors. Fields virology. 5th edition. Philadelphia, PA: Lippincott William & Wilkins; 2007. pp. 1101–1152.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

