High motility of triple-negative breast cancer cells is due to repression of plakoglobin gene by metastasis modulator protein SLUG
- PMID: 22496452
- PMCID: PMC3365985
- DOI: 10.1074/jbc.M112.345728
High motility of triple-negative breast cancer cells is due to repression of plakoglobin gene by metastasis modulator protein SLUG
Abstract
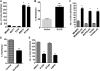
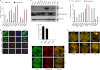
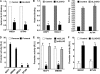
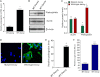
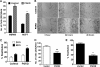
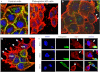
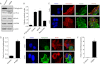
One of highly pathogenic breast cancer cell types are the triple negative (negative in the expression of estrogen, progesterone, and ERBB2 receptors) breast cancer cells. These cells are highly motile and metastatic and are characterized by high levels of the metastasis regulator protein SLUG. Using isogenic breast cancer cell systems we have shown here that high motility of these cells is directly correlated with the levels of the SLUG in these cells. Because epithelial/mesenchymal cell motility is known to be negatively regulated by the catenin protein plakoglobin, we postulated that the transcriptional repressor protein SLUG increases the motility of the aggressive breast cancer cells through the knockdown of the transcription of the plakoglobin gene. We found that SLUG inhibits the expression of plakoglobin gene directly in these cells. Overexpression of SLUG in the SLUG-deficient cancer cells significantly decreased the levels of mRNA and protein of plakoglobin. On the contrary, knockdown of SLUG in SLUG-high cancer cells elevated the levels of plakoglobin. Blocking of SLUG function with a double-stranded DNA decoy that competes with the E2-box binding of SLUG also increased the levels of plakoglobin mRNA, protein, and promoter activity in the SLUG-high triple negative breast cancer cells. Overexpression of SLUG in the SLUG-deficient cells elevated the motility of these cells. Knockdown of plakoglobin in these low motility non-invasive breast cancer cells rearranged the actin filaments and increased the motility of these cells. Forced expression of plakoglobin in SLUG-high cells had the reverse effects on cellular motility. This study thus implicates SLUG-induced repression of plakoglobin as a motility determinant in highly disseminating breast cancer.
Figures



Similar articles
-
Plakoglobin represses SATB1 expression and decreases in vitro proliferation, migration and invasion.PLoS One. 2013 Nov 8;8(11):e78388. doi: 10.1371/journal.pone.0078388. eCollection 2013. PLoS One. 2013. PMID: 24260116 Free PMC article.
-
SLUG-induced elevation of D1 cyclin in breast cancer cells through the inhibition of its ubiquitination.J Biol Chem. 2011 Jan 7;286(1):469-79. doi: 10.1074/jbc.M110.164384. Epub 2010 Nov 2. J Biol Chem. 2011. PMID: 21044962 Free PMC article.
-
DNMT3A and DNMT3B mediate autocrine hGH repression of plakoglobin gene transcription and consequent phenotypic conversion of mammary carcinoma cells.Oncogene. 2008 Apr 17;27(18):2602-12. doi: 10.1038/sj.onc.1210917. Epub 2007 Nov 12. Oncogene. 2008. PMID: 17998942
-
Regulation of BRCA2 gene expression by the SLUG repressor protein in human breast cells.J Biol Chem. 2005 Apr 29;280(17):17163-71. doi: 10.1074/jbc.M501375200. Epub 2005 Feb 24. J Biol Chem. 2005. PMID: 15734731 Free PMC article.
-
Circulating tumor cell clusters-associated gene plakoglobin and breast cancer survival.Breast Cancer Res Treat. 2015 Jun;151(3):491-500. doi: 10.1007/s10549-015-3416-1. Epub 2015 May 10. Breast Cancer Res Treat. 2015. PMID: 25957595 Review.
Cited by
-
XAF1 promotes colorectal cancer metastasis via VCP-RNF114-JUP axis.J Cell Biol. 2024 Feb 5;223(2):e202303015. doi: 10.1083/jcb.202303015. Epub 2023 Dec 14. J Cell Biol. 2024. PMID: 38095639 Free PMC article.
-
Implications of PPPDE1 expression in the distribution of plakoglobin and β-catenin in pancreatic ductal adenocarcinoma.Oncol Lett. 2014 Sep;8(3):1229-1233. doi: 10.3892/ol.2014.2279. Epub 2014 Jun 24. Oncol Lett. 2014. PMID: 25120694 Free PMC article.
-
The role of the redox/miR-6855-3p/PRDX5A axis in reversing SLUG-mediated BRCA2 silencing in breast cancer cells.Cell Commun Signal. 2020 Jan 27;18(1):15. doi: 10.1186/s12964-019-0493-5. Cell Commun Signal. 2020. PMID: 31987042 Free PMC article.
-
Activation of KRAS promotes the mesenchymal features of basal-type breast cancer.Exp Mol Med. 2015 Jan 30;47(1):e137. doi: 10.1038/emm.2014.99. Exp Mol Med. 2015. PMID: 25633745 Free PMC article.
-
Cytotoxicity, Post-Treatment Recovery, and Selectivity Analysis of Naturally Occurring Podophyllotoxins from Bursera fagaroides var. fagaroides on Breast Cancer Cell Lines.Molecules. 2016 Aug 4;21(8):1013. doi: 10.3390/molecules21081013. Molecules. 2016. PMID: 27527135 Free PMC article.
References
-
- Huo D., Ikpatt F., Khramtsov A., Dangou J. M., Nanda R., Dignam J., Zhang B., Grushko T., Zhang C., Oluwasola O., Malaka D., Malami S., Odetunde A., Adeoye A. O., Iyare F., Falusi A., Perou C. M., Olopade O. I. (2009) Population differences in breast cancer. Survey in indigenous African women reveals over-representation of triple-negative breast cancer. J. Clin. Oncol. 27, 4515–4521 - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

