Alternative splicing of CHEK2 and codeletion with NF2 promote chromosomal instability in meningioma
- PMID: 22355270
- PMCID: PMC3281938
- DOI: 10.1593/neo.111574
Alternative splicing of CHEK2 and codeletion with NF2 promote chromosomal instability in meningioma
Abstract
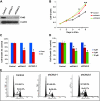
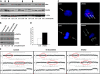
Mutations of the NF2 gene on chromosome 22q are thought to initiate tumorigenesis in nearly 50% of meningiomas, and 22q deletion is the earliest and most frequent large-scale chromosomal abnormality observed in these tumors. In aggressive meningiomas, 22q deletions are generally accompanied by the presence of large-scale segmental abnormalities involving other chromosomes, but the reasons for this association are unknown. We find that large-scale chromosomal alterations accumulate during meningioma progression primarily in tumors harboring 22q deletions, suggesting 22q-associated chromosomal instability. Here we show frequent codeletion of the DNA repair and tumor suppressor gene, CHEK2, in combination with NF2 on chromosome 22q in a majority of aggressive meningiomas. In addition, tumor-specific splicing of CHEK2 in meningioma leads to decreased functional Chk2 protein expression. We show that enforced Chk2 knockdown in meningioma cells decreases DNA repair. Furthermore, Chk2 depletion increases centrosome amplification, thereby promoting chromosomal instability. Taken together, these data indicate that alternative splicing and frequent codeletion of CHEK2 and NF2 contribute to the genomic instability and associated development of aggressive biologic behavior in meningiomas.
Figures



Similar articles
-
NF2 mutations in secretory and other rare variants of meningiomas.Brain Pathol. 2006 Jan;16(1):15-9. doi: 10.1111/j.1750-3639.2006.tb00557.x. Brain Pathol. 2006. PMID: 16612978 Free PMC article.
-
Comprehensive genetic and epigenetic analysis of sporadic meningioma for macro-mutations on 22q and micro-mutations within the NF2 locus.BMC Genomics. 2007 Jan 12;8:16. doi: 10.1186/1471-2164-8-16. BMC Genomics. 2007. PMID: 17222329 Free PMC article.
-
Combined molecular genetic studies of chromosome 22q and the neurofibromatosis type 2 gene in central nervous system tumors.Neurosurgery. 1995 Oct;37(4):764-73. doi: 10.1227/00006123-199510000-00022. Neurosurgery. 1995. PMID: 8559307
-
[The molecular genetics of meningiomas and genotypic/phenotypic correlations].Rev Neurol (Paris). 2003 Sep;159(8-9):727-38. Rev Neurol (Paris). 2003. PMID: 13679715 Review. French.
-
Pathological classification and molecular genetics of meningiomas.J Neurooncol. 2010 Sep;99(3):379-91. doi: 10.1007/s11060-010-0342-2. Epub 2010 Sep 1. J Neurooncol. 2010. PMID: 20809251 Review.
Cited by
-
Cancer subclonal genetic architecture as a key to personalized medicine.Neoplasia. 2013 Dec;15(12):1410-20. doi: 10.1593/neo.131972. Neoplasia. 2013. PMID: 24403863 Free PMC article.
-
Domestic Animal Models of Central Nervous System Tumors: Focus on Meningiomas.Life (Basel). 2023 Nov 30;13(12):2284. doi: 10.3390/life13122284. Life (Basel). 2023. PMID: 38137885 Free PMC article. Review.
-
MicroRNA-191 promotes osteosarcoma cells proliferation by targeting checkpoint kinase 2.Tumour Biol. 2015 Aug;36(8):6095-101. doi: 10.1007/s13277-015-3290-9. Epub 2015 Mar 14. Tumour Biol. 2015. PMID: 25773391
-
Chromosome 22q12.1 microdeletions: confirmation of the MN1 gene as a candidate gene for cleft palate.Eur J Hum Genet. 2016 Jan;24(1):51-8. doi: 10.1038/ejhg.2015.65. Epub 2015 May 6. Eur J Hum Genet. 2016. PMID: 25944382 Free PMC article.
-
Genomic sequencing of meningiomas identifies oncogenic SMO and AKT1 mutations.Nat Genet. 2013 Mar;45(3):285-9. doi: 10.1038/ng.2526. Epub 2013 Jan 20. Nat Genet. 2013. PMID: 23334667 Free PMC article.
References
-
- Arslantas A, Artan S, Oner U, Durmaz R, Muslumanoglu H, Atasoy MA, Basaran N, Tel E. Comparative genomic hybridization analysis of genomic alterations in benign, atypical and anaplastic meningiomas. Acta Neurol Belg. 2002;102:53–62. - PubMed
-
- Goutagny S, Yang HW, Zucman-Rossi J, Chan J, Dreyfuss JM, Park PJ, Black PM, Giovannini M, Carroll RS, Kalamarides M. Genomic profiling reveals alternative genetic pathways of meningioma malignant progression dependent on the underlying NF2 status. Clin Cancer Res. 2010;16:4155–4164. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous
