PP4 dephosphorylates Maf1 to couple multiple stress conditions to RNA polymerase III repression
- PMID: 22333918
- PMCID: PMC3321174
- DOI: 10.1038/emboj.2011.501
PP4 dephosphorylates Maf1 to couple multiple stress conditions to RNA polymerase III repression
Abstract
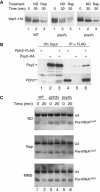
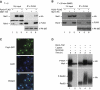
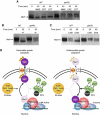
Maf1 is the 'master' repressor of RNA polymerase III (Pol III) transcription in yeast, and is conserved in eukaryotes. Maf1 is a phospho-integrator, with unfavourable growth conditions leading to rapid Maf1 dephosphorylation, nuclear accumulation, binding to RNA Pol III at Pol III genes and transcriptional repression. Here, we establish the protein phosphatase 4 (PP4) complex as the main Maf1 phosphatase, and define the involved catalytic (Pph3), scaffold (Psy2) and regulatory subunits (Rrd1, Tip41), as well as uninvolved subunits (Psy4, Rrd2). Multiple approaches support a central role for PP4 in Maf1 dephosphorylation, Maf1 nuclear localization and the rapid repression of Pol III in the nucleus. PP4 action is likely direct, as a portion of PP4 co-precipitates with Maf1, and purified PP4 dephosphorylates Maf1 in vitro. Furthermore, Pph3 mediates (either largely or fully) rapid Maf1 dephosphorylation in response to diverse stresses, suggesting PP4 plays a key role in the integration of cell nutrition and stress conditions by Maf1 to enable Pol III regulation.
Conflict of interest statement
The authors declare that they have no conflict of interest.
Figures




Similar articles
-
Maf1 phosphorylation is regulated through the action of prefoldin-like Bud27 on PP4 phosphatase in Saccharomyces cerevisiae.Nucleic Acids Res. 2024 Jul 8;52(12):7081-7095. doi: 10.1093/nar/gkae414. Nucleic Acids Res. 2024. PMID: 38864693 Free PMC article.
-
General repression of RNA polymerase III transcription is triggered by protein phosphatase type 2A-mediated dephosphorylation of Maf1.Mol Cell. 2006 Jun 9;22(5):623-32. doi: 10.1016/j.molcel.2006.04.008. Mol Cell. 2006. PMID: 16762835
-
Dephosphorylation and genome-wide association of Maf1 with Pol III-transcribed genes during repression.Mol Cell. 2006 Jun 9;22(5):633-44. doi: 10.1016/j.molcel.2006.04.009. Mol Cell. 2006. PMID: 16762836 Free PMC article.
-
Integration of nutritional and stress signaling pathways by Maf1.Trends Biochem Sci. 2007 Feb;32(2):51-3. doi: 10.1016/j.tibs.2006.12.001. Epub 2006 Dec 14. Trends Biochem Sci. 2007. PMID: 17174096 Review.
-
Maf1, a general negative regulator of RNA polymerase III in yeast.Biochim Biophys Acta. 2013 Mar-Apr;1829(3-4):376-84. doi: 10.1016/j.bbagrm.2012.11.004. Epub 2012 Nov 28. Biochim Biophys Acta. 2013. PMID: 23201230 Review.
Cited by
-
A Consensus Binding Motif for the PP4 Protein Phosphatase.Mol Cell. 2019 Dec 19;76(6):953-964.e6. doi: 10.1016/j.molcel.2019.08.029. Epub 2019 Oct 1. Mol Cell. 2019. PMID: 31585692 Free PMC article.
-
Glycolytic flux in Saccharomyces cerevisiae is dependent on RNA polymerase III and its negative regulator Maf1.Biochem J. 2019 Apr 4;476(7):1053-1082. doi: 10.1042/BCJ20180701. Biochem J. 2019. PMID: 30885983 Free PMC article.
-
Functional roles of protein phosphatase 4 in multiple aspects of cellular physiology: a friend and a foe.BMB Rep. 2020 Apr;53(4):181-190. doi: 10.5483/BMBRep.2020.53.4.019. BMB Rep. 2020. PMID: 32192570 Free PMC article. Review.
-
Codon-biased translation can be regulated by wobble-base tRNA modification systems during cellular stress responses.RNA Biol. 2015;12(6):603-14. doi: 10.1080/15476286.2015.1031947. RNA Biol. 2015. PMID: 25892531 Free PMC article. Review.
-
Ras/ERK-signalling promotes tRNA synthesis and growth via the RNA polymerase III repressor Maf1 in Drosophila.PLoS Genet. 2018 Feb 5;14(2):e1007202. doi: 10.1371/journal.pgen.1007202. eCollection 2018 Feb. PLoS Genet. 2018. PMID: 29401457 Free PMC article.
References
-
- Boguta M, Czerska K, Zoladek T (1997) Mutation in a new gene MAF1 affects tRNA suppressor efficiency in Saccharomyces cerevisiae. Gene 185: 291–296 - PubMed
-
- Cohen PT, Philp A, Vazquez-Martin C (2005) Protein phosphatase 4—from obscurity to vital functions. FEBS Lett 579: 3278–3286 - PubMed
-
- Collins SR, Kemmeren P, Zhao XC, Greenblatt JF, Spencer F, Holstege FC, Weissman JS, Krogan NJ (2007) Toward a comprehensive atlas of the physical interactome of Saccharomyces cerevisiae. Mol Cell Proteomics 6: 439–450 - PubMed
-
- Desai N, Lee J, Upadhya R, Chu Y, Moir RD, Willis IM (2005) Two steps in Maf1-dependent repression of transcription by RNA polymerase III. J Biol Chem 280: 6455–6462 - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

