The RBP-Jκ binding sites within the RTA promoter regulate KSHV latent infection and cell proliferation
- PMID: 22253595
- PMCID: PMC3257303
- DOI: 10.1371/journal.ppat.1002479
The RBP-Jκ binding sites within the RTA promoter regulate KSHV latent infection and cell proliferation
Abstract
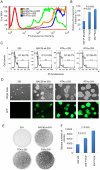
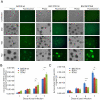
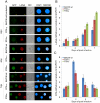
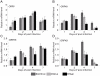
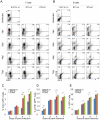
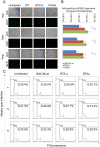
Kaposi's sarcoma-associated herpesvirus (KSHV) is tightly linked to at least two lymphoproliferative disorders, primary effusion lymphoma (PEL) and multicentric Castleman's disease (MCD). However, the development of KSHV-mediated lymphoproliferative disease is not fully understood. Here, we generated two recombinant KSHV viruses deleted for the first RBP-Jκ binding site (RTA(1st)) and all three RBP-Jκ binding sites (RTA(all)) within the RTA promoter. Our results showed that RTA(1st) and RTA(all) recombinant viruses possess increased viral latency and a decreased capability for lytic replication in HEK 293 cells, enhancing colony formation and proliferation of infected cells. Furthermore, recombinant RTA(1st) and RTA(all) viruses showed greater infectivity in human peripheral blood mononuclear cells (PBMCs) relative to wt KSHV. Interestingly, KSHV BAC36 wt, RTA(1st) and RTA(all) recombinant viruses infected both T and B cells and all three viruses efficiently infected T and B cells in a time-dependent manner early after infection. Also, the capability of both RTA(1st) and RTA(all) recombinant viruses to infect CD19+ B cells was significantly enhanced. Surprisingly, RTA(1st) and RTA(all) recombinant viruses showed greater infectivity for CD3+ T cells up to 7 days. Furthermore, studies in Telomerase-immortalized human umbilical vein endothelial (TIVE) cells infected with KSHV corroborated our data that RTA(1st) and RTA(all) recombinant viruses have enhanced ability to persist in latently infected cells with increased proliferation. These recombinant viruses now provide a model to explore early stages of primary infection in human PBMCs and development of KSHV-associated lymphoproliferative diseases.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures



Similar articles
-
The single RBP-Jkappa site within the LANA promoter is crucial for establishing Kaposi's sarcoma-associated herpesvirus latency during primary infection.J Virol. 2011 Jul;85(13):6148-61. doi: 10.1128/JVI.02608-10. Epub 2011 Apr 20. J Virol. 2011. PMID: 21507979 Free PMC article.
-
Genome-Wide Identification of Direct RTA Targets Reveals Key Host Factors for Kaposi's Sarcoma-Associated Herpesvirus Lytic Reactivation.J Virol. 2019 Feb 19;93(5):e01978-18. doi: 10.1128/JVI.01978-18. Print 2019 Mar 1. J Virol. 2019. PMID: 30541837 Free PMC article.
-
NF-kappaB serves as a cellular sensor of Kaposi's sarcoma-associated herpesvirus latency and negatively regulates K-Rta by antagonizing the RBP-Jkappa coactivator.J Virol. 2009 May;83(9):4435-46. doi: 10.1128/JVI.01999-08. Epub 2009 Feb 25. J Virol. 2009. PMID: 19244329 Free PMC article.
-
Inhibition of KAP1 enhances hypoxia-induced Kaposi's sarcoma-associated herpesvirus reactivation through RBP-Jκ.J Virol. 2014 Jun;88(12):6873-84. doi: 10.1128/JVI.00283-14. Epub 2014 Apr 2. J Virol. 2014. PMID: 24696491 Free PMC article.
-
Epigenetic regulation of Kaposi's sarcoma-associated herpesvirus replication.Semin Cancer Biol. 2009 Jun;19(3):153-7. doi: 10.1016/j.semcancer.2009.02.010. Epub 2009 Feb 21. Semin Cancer Biol. 2009. PMID: 19429478 Free PMC article. Review.
Cited by
-
Kaposi's sarcoma-associated herpesvirus-encoded LANA contributes to viral latent replication by activating phosphorylation of survivin.J Virol. 2014 Apr;88(8):4204-17. doi: 10.1128/JVI.03855-13. Epub 2014 Jan 29. J Virol. 2014. PMID: 24478433 Free PMC article.
-
Constitutive Activation of Interleukin-13/STAT6 Contributes to Kaposi's Sarcoma-Associated Herpesvirus-Related Primary Effusion Lymphoma Cell Proliferation and Survival.J Virol. 2015 Oct;89(20):10416-26. doi: 10.1128/JVI.01525-15. Epub 2015 Aug 5. J Virol. 2015. PMID: 26246572 Free PMC article.
-
Abortive lytic reactivation of KSHV in CBF1/CSL deficient human B cell lines.PLoS Pathog. 2013;9(5):e1003336. doi: 10.1371/journal.ppat.1003336. Epub 2013 May 16. PLoS Pathog. 2013. PMID: 23696732 Free PMC article.
-
An EBV recombinant deleted for residues 130-159 in EBNA3C can deregulate p53/Mdm2 and Cyclin D1/CDK6 which results in apoptosis and reduced cell proliferation.Oncotarget. 2016 Apr 5;7(14):18116-34. doi: 10.18632/oncotarget.7502. Oncotarget. 2016. PMID: 26908453 Free PMC article.
-
Polyamine biosynthesis and eIF5A hypusination are modulated by the DNA tumor virus KSHV and promote KSHV viral infection.PLoS Pathog. 2022 Apr 29;18(4):e1010503. doi: 10.1371/journal.ppat.1010503. eCollection 2022 Apr. PLoS Pathog. 2022. PMID: 35486659 Free PMC article.
References
-
- Cesarman E, Chang Y, Moore PS, Said JW, Knowles DM. Kaposi's sarcoma-associated herpesvirus-like DNA sequences in AIDS-related body-cavity-based lymphomas. N Engl J Med. 1995;332:1186–1191. - PubMed
-
- Soulier J, Grollet L, Oksenhendler E, Cacoub P, Cazals-Hatem D, et al. Kaposi's sarcoma-associated herpesvirus-like DNA sequences in multicentric Castleman's disease. Blood. 1995;86:1276–1280. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

