Ric-8 proteins are molecular chaperones that direct nascent G protein α subunit membrane association
- PMID: 22114146
- PMCID: PMC3870195
- DOI: 10.1126/scisignal.2002223
Ric-8 proteins are molecular chaperones that direct nascent G protein α subunit membrane association
Abstract
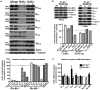
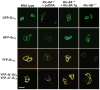
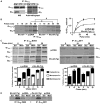
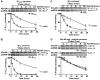
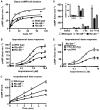
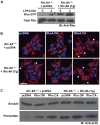
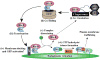
Ric-8A (resistance to inhibitors of cholinesterase 8A) and Ric-8B are guanine nucleotide exchange factors that enhance different heterotrimeric guanine nucleotide-binding protein (G protein) signaling pathways by unknown mechanisms. Because transgenic disruption of Ric-8A or Ric-8B in mice caused early embryonic lethality, we derived viable Ric-8A- or Ric-8B-deleted embryonic stem (ES) cell lines from blastocysts of these mice. We observed pleiotropic G protein signaling defects in Ric-8A(-/-) ES cells, which resulted from reduced steady-state amounts of Gα(i), Gα(q), and Gα(13) proteins to <5% of those of wild-type cells. The amounts of Gα(s) and total Gβ protein were partially reduced in Ric-8A(-/-) cells compared to those in wild-type cells, and only the amount of Gα(s) was reduced substantially in Ric-8B(-/-) cells. The abundances of mRNAs encoding the G protein α subunits were largely unchanged by loss of Ric-8A or Ric-8B. The plasma membrane residence of G proteins persisted in the absence of Ric-8 but was markedly reduced compared to that in wild-type cells. Endogenous Gα(i) and Gα(q) were efficiently translated in Ric-8A(-/-) cells but integrated into endomembranes poorly; however, the reduced amounts of G protein α subunits that reached the membrane still bound to nascent Gβγ. Finally, Gα(i), Gα(q), and Gβ(1) proteins exhibited accelerated rates of degradation in Ric-8A(-/-) cells compared to those in wild-type cells. Together, these data suggest that Ric-8 proteins are molecular chaperones required for the initial association of nascent Gα subunits with cellular membranes.
Conflict of interest statement
Figures

Similar articles
-
Ric-8B is a GTP-dependent G protein alphas guanine nucleotide exchange factor.J Biol Chem. 2011 Jun 3;286(22):19932-42. doi: 10.1074/jbc.M110.163675. Epub 2011 Apr 5. J Biol Chem. 2011. PMID: 21467038 Free PMC article.
-
Molecular chaperoning function of Ric-8 is to fold nascent heterotrimeric G protein α subunits.Proc Natl Acad Sci U S A. 2013 Mar 5;110(10):3794-9. doi: 10.1073/pnas.1220943110. Epub 2013 Feb 19. Proc Natl Acad Sci U S A. 2013. PMID: 23431197 Free PMC article.
-
Dual phosphorylation of Ric-8A enhances its ability to mediate G protein α subunit folding and to stimulate guanine nucleotide exchange.Sci Signal. 2018 May 29;11(532):eaap8113. doi: 10.1126/scisignal.aap8113. Sci Signal. 2018. PMID: 29844055 Free PMC article.
-
Ric-8 regulation of heterotrimeric G proteins.J Recept Signal Transduct Res. 2013 Jun;33(3):139-43. doi: 10.3109/10799893.2013.763828. Epub 2013 Feb 6. J Recept Signal Transduct Res. 2013. PMID: 23384070 Free PMC article. Review.
-
The G protein α chaperone Ric-8 as a potential therapeutic target.Mol Pharmacol. 2015 Jan;87(1):52-63. doi: 10.1124/mol.114.094664. Epub 2014 Oct 15. Mol Pharmacol. 2015. PMID: 25319541 Free PMC article. Review.
Cited by
-
Expression Pattern and Localization Dynamics of Guanine Nucleotide Exchange Factor RIC8 during Mouse Oogenesis.PLoS One. 2015 Jun 10;10(6):e0129131. doi: 10.1371/journal.pone.0129131. eCollection 2015. PLoS One. 2015. PMID: 26062014 Free PMC article.
-
Production of Phosphorylated Ric-8A proteins using protein kinase CK2.Protein Expr Purif. 2019 Feb;154:98-103. doi: 10.1016/j.pep.2018.10.002. Epub 2018 Oct 2. Protein Expr Purif. 2019. PMID: 30290220 Free PMC article.
-
Chaperoning G protein-coupled receptors: from cell biology to therapeutics.Endocr Rev. 2014 Aug;35(4):602-47. doi: 10.1210/er.2013-1121. Epub 2014 Mar 24. Endocr Rev. 2014. PMID: 24661201 Free PMC article. Review.
-
Ablation of RIC8A function in mouse neurons leads to a severe neuromuscular phenotype and postnatal death.PLoS One. 2013 Aug 16;8(8):e74031. doi: 10.1371/journal.pone.0074031. eCollection 2013. PLoS One. 2013. PMID: 23977396 Free PMC article.
-
Canonical and noncanonical g-protein signaling helps coordinate actin dynamics to promote macrophage phagocytosis of zymosan.Mol Cell Biol. 2014 Nov 15;34(22):4186-99. doi: 10.1128/MCB.00325-14. Epub 2014 Sep 15. Mol Cell Biol. 2014. PMID: 25225330 Free PMC article.
References
-
- Tall GG, Krumins AM, Gilman AG. Mammalian Ric-8A (synembryn) is a heterotrimeric Gα protein guanine nucleotide exchange factor. J Biol Chem. 2003;278:8356–8362. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

