PPARα-Sirt1 complex mediates cardiac hypertrophy and failure through suppression of the ERR transcriptional pathway
- PMID: 22055503
- PMCID: PMC3217210
- DOI: 10.1016/j.cmet.2011.10.001
PPARα-Sirt1 complex mediates cardiac hypertrophy and failure through suppression of the ERR transcriptional pathway
Abstract
High energy production in mitochondria is essential for maintaining cardiac contraction in the heart. Genes regulating mitochondrial function are commonly downregulated during heart failure. Here we show that both PPARα and Sirt1 are upregulated by pressure overload in the heart. Haploinsufficiency of either PPARα or Sirt1 attenuated pressure overload-induced cardiac hypertrophy and failure, whereas simultaneous upregulation of PPARα and Sirt1 exacerbated the cardiac dysfunction. PPARα and Sirt1 coordinately suppressed genes involved in mitochondrial function that are regulated by estrogen-related receptors (ERRs). PPARα bound and recruited Sirt1 to the ERR response element (ERRE), thereby suppressing ERR target genes in an RXR-independent manner. Downregulation of ERR target genes was also observed during fasting, and this appeared to be an adaptive response of the heart. These results suggest that suppression of the ERR transcriptional pathway by PPARα/Sirt1, a physiological fasting response, is involved in the progression of heart failure by promoting mitochondrial dysfunction.
Copyright © 2011 Elsevier Inc. All rights reserved.
Figures

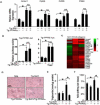
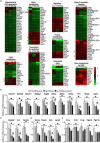
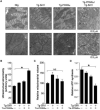


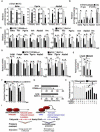
Similar articles
-
Suppression of ERR targets by a PPARα/Sirt1 complex in the failing heart.Cell Cycle. 2012 Mar 1;11(5):856-64. doi: 10.4161/cc.11.5.19210. Epub 2012 Mar 1. Cell Cycle. 2012. PMID: 22333581 Free PMC article.
-
Peroxisome Proliferator Activated Receptor-α Association With Silent Information Regulator 1 Suppresses Cardiac Fatty Acid Metabolism in the Failing Heart.Circ Heart Fail. 2015 Nov;8(6):1123-32. doi: 10.1161/CIRCHEARTFAILURE.115.002216. Epub 2015 Oct 6. Circ Heart Fail. 2015. PMID: 26443578 Free PMC article.
-
Sirt1 acts in association with PPARα to protect the heart from hypertrophy, metabolic dysregulation, and inflammation.Cardiovasc Res. 2011 May 1;90(2):276-84. doi: 10.1093/cvr/cvq376. Epub 2010 Nov 29. Cardiovasc Res. 2011. PMID: 21115502
-
Metabolic reprogramming via PPARα signaling in cardiac hypertrophy and failure: From metabolomics to epigenetics.Am J Physiol Heart Circ Physiol. 2017 Sep 1;313(3):H584-H596. doi: 10.1152/ajpheart.00103.2017. Epub 2017 Jun 23. Am J Physiol Heart Circ Physiol. 2017. PMID: 28646024 Free PMC article. Review.
-
Mitochondria in cardiac hypertrophy and heart failure.J Mol Cell Cardiol. 2013 Feb;55:31-41. doi: 10.1016/j.yjmcc.2012.09.002. Epub 2012 Sep 13. J Mol Cell Cardiol. 2013. PMID: 22982369 Free PMC article. Review.
Cited by
-
Overview of pyridine nucleotides review series.Circ Res. 2012 Aug 17;111(5):604-10. doi: 10.1161/CIRCRESAHA.111.247924. Circ Res. 2012. PMID: 22904040 Free PMC article. Review.
-
The Role of Posttranslational Modification and Mitochondrial Quality Control in Cardiovascular Diseases.Oxid Med Cell Longev. 2021 Feb 18;2021:6635836. doi: 10.1155/2021/6635836. eCollection 2021. Oxid Med Cell Longev. 2021. Retraction in: Oxid Med Cell Longev. 2023 Oct 11;2023:9821720. doi: 10.1155/2023/9821720. PMID: 33680284 Free PMC article. Retracted. Review.
-
Expression and Function of PPARs in Placenta.PPAR Res. 2013;2013:256508. doi: 10.1155/2013/256508. Epub 2013 Feb 12. PPAR Res. 2013. PMID: 23476631 Free PMC article.
-
NAD metabolism and heart failure: Mechanisms and therapeutic potentials.J Mol Cell Cardiol. 2024 Oct;195:45-54. doi: 10.1016/j.yjmcc.2024.07.008. Epub 2024 Aug 3. J Mol Cell Cardiol. 2024. PMID: 39096536 Review.
-
Circulating anti-geronic factors from heterochonic parabionts promote vascular rejuvenation in aged mice: transcriptional footprint of mitochondrial protection, attenuation of oxidative stress, and rescue of endothelial function by young blood.Geroscience. 2020 Apr;42(2):727-748. doi: 10.1007/s11357-020-00180-6. Epub 2020 Mar 15. Geroscience. 2020. PMID: 32172434 Free PMC article.
References
-
- Alcendor RR, Gao S, Zhai P, Zablocki D, Holle E, Yu X, Tian B, Wagner T, Vatner SF, Sadoshima J. Sirt1 regulates aging and resistance to oxidative stress in the heart. Circ Res. 2007;100:1512–1521. - PubMed
-
- Bordone L, Guarente L. Calorie restriction, SIRT1 and metabolism: understanding longevity. Nat Rev Mol Cell Biol. 2005;6:298–305. - PubMed
-
- Dufour CR, Wilson BJ, Huss JM, Kelly DP, Alaynick WA, Downes M, Evans RM, Blanchette M, Giguere V. Genome-wide orchestration of cardiac functions by the orphan nuclear receptors ERRalpha and gamma. Cell Metab. 2007;5:345–356. - PubMed
-
- Giguere V. Transcriptional control of energy homeostasis by the estrogen-related receptors. Endocr Rev. 2008;29:677–696. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- P01 AG027211/AG/NIA NIH HHS/United States
- R01 HL067724-12/HL/NHLBI NIH HHS/United States
- R01 HL067724-05/HL/NHLBI NIH HHS/United States
- R01 AG023039-03/AG/NIA NIH HHS/United States
- HL91469/HL/NHLBI NIH HHS/United States
- R01 HL067724-06/HL/NHLBI NIH HHS/United States
- P01 HL069020/HL/NHLBI NIH HHS/United States
- R21 HL098802/HL/NHLBI NIH HHS/United States
- R01 HL067724/HL/NHLBI NIH HHS/United States
- P01 AG027211-05/AG/NIA NIH HHS/United States
- R01 HL067724-03/HL/NHLBI NIH HHS/United States
- R01 AG028787/AG/NIA NIH HHS/United States
- HL67724/HL/NHLBI NIH HHS/United States
- R01 HL102738-01/HL/NHLBI NIH HHS/United States
- AG27211/AG/NIA NIH HHS/United States
- R01 HL102738/HL/NHLBI NIH HHS/United States
- R01 HL067727-01/HL/NHLBI NIH HHS/United States
- R01 AG023039-04/AG/NIA NIH HHS/United States
- AG23039/AG/NIA NIH HHS/United States
- R01 AG023039-08/AG/NIA NIH HHS/United States
- P01 AG027211-02/AG/NIA NIH HHS/United States
- R01 HL091469/HL/NHLBI NIH HHS/United States
- R01 AG028787-01/AG/NIA NIH HHS/United States
- R01 HL091469-02W1/HL/NHLBI NIH HHS/United States
- R01 HL067724-02/HL/NHLBI NIH HHS/United States
- R01 HL067724-10/HL/NHLBI NIH HHS/United States
- R01 HL091469-03/HL/NHLBI NIH HHS/United States
- R01 HL067724-01/HL/NHLBI NIH HHS/United States
- P01 AG027211-02S1/AG/NIA NIH HHS/United States
- R01 AG023039-02/AG/NIA NIH HHS/United States
- R01 AG023039-07/AG/NIA NIH HHS/United States
- R01 HL091469-01/HL/NHLBI NIH HHS/United States
- R01 HL102738-02/HL/NHLBI NIH HHS/United States
- HL69020/HL/NHLBI NIH HHS/United States
- R01 HL067724-08/HL/NHLBI NIH HHS/United States
- R01 HL102738-03/HL/NHLBI NIH HHS/United States
- R01 HL067724-09/HL/NHLBI NIH HHS/United States
- R01 AG023039-06S1/AG/NIA NIH HHS/United States
- R01 HL091469-05/HL/NHLBI NIH HHS/United States
- P01 AG027211-04/AG/NIA NIH HHS/United States
- P01 AG027211-01A1/AG/NIA NIH HHS/United States
- R01 AG023039-05/AG/NIA NIH HHS/United States
- R01 HL091469-02/HL/NHLBI NIH HHS/United States
- P01 AG027211-03/AG/NIA NIH HHS/United States
- HL102738/HL/NHLBI NIH HHS/United States
- R01 HL067724-04/HL/NHLBI NIH HHS/United States
- R01 HL112330/HL/NHLBI NIH HHS/United States
- R01 HL067724-11/HL/NHLBI NIH HHS/United States
- R01 AG023039-01/AG/NIA NIH HHS/United States
- R01 HL067724-07/HL/NHLBI NIH HHS/United States
- R01 AG023039/AG/NIA NIH HHS/United States
- R01 AG023039-09/AG/NIA NIH HHS/United States
- R01 HL091469-04/HL/NHLBI NIH HHS/United States
- R01 AG023039-06/AG/NIA NIH HHS/United States
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases

