Cleavage and activation of the severe acute respiratory syndrome coronavirus spike protein by human airway trypsin-like protease
- PMID: 21994442
- PMCID: PMC3233180
- DOI: 10.1128/JVI.05300-11
Cleavage and activation of the severe acute respiratory syndrome coronavirus spike protein by human airway trypsin-like protease
Abstract
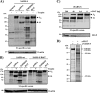
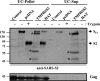
The highly pathogenic severe acute respiratory syndrome coronavirus (SARS-CoV) poses a constant threat to human health. The viral spike protein (SARS-S) mediates host cell entry and is a potential target for antiviral intervention. Activation of SARS-S by host cell proteases is essential for SARS-CoV infectivity but remains incompletely understood. Here, we analyzed the role of the type II transmembrane serine proteases (TTSPs) human airway trypsin-like protease (HAT) and transmembrane protease, serine 2 (TMPRSS2), in SARS-S activation. We found that HAT activates SARS-S in the context of surrogate systems and authentic SARS-CoV infection and is coexpressed with the viral receptor angiotensin-converting enzyme 2 (ACE2) in bronchial epithelial cells and pneumocytes. HAT cleaved SARS-S at R667, as determined by mutagenesis and mass spectrometry, and activated SARS-S for cell-cell fusion in cis and trans, while the related pulmonary protease TMPRSS2 cleaved SARS-S at multiple sites and activated SARS-S only in trans. However, TMPRSS2 but not HAT expression rendered SARS-S-driven virus-cell fusion independent of cathepsin activity, indicating that HAT and TMPRSS2 activate SARS-S differentially. Collectively, our results show that HAT cleaves and activates SARS-S and might support viral spread in patients.
Figures

Similar articles
-
TMPRSS2 and ADAM17 cleave ACE2 differentially and only proteolysis by TMPRSS2 augments entry driven by the severe acute respiratory syndrome coronavirus spike protein.J Virol. 2014 Jan;88(2):1293-307. doi: 10.1128/JVI.02202-13. Epub 2013 Nov 13. J Virol. 2014. PMID: 24227843 Free PMC article.
-
Evidence that TMPRSS2 activates the severe acute respiratory syndrome coronavirus spike protein for membrane fusion and reduces viral control by the humoral immune response.J Virol. 2011 May;85(9):4122-34. doi: 10.1128/JVI.02232-10. Epub 2011 Feb 16. J Virol. 2011. PMID: 21325420 Free PMC article.
-
Efficient activation of the severe acute respiratory syndrome coronavirus spike protein by the transmembrane protease TMPRSS2.J Virol. 2010 Dec;84(24):12658-64. doi: 10.1128/JVI.01542-10. Epub 2010 Oct 6. J Virol. 2010. PMID: 20926566 Free PMC article.
-
Proteolytic activation of the SARS-coronavirus spike protein: cutting enzymes at the cutting edge of antiviral research.Antiviral Res. 2013 Dec;100(3):605-14. doi: 10.1016/j.antiviral.2013.09.028. Epub 2013 Oct 8. Antiviral Res. 2013. PMID: 24121034 Free PMC article. Review.
-
Severe acute respiratory syndrome coronavirus entry as a target of antiviral therapies.Antivir Ther. 2007;12(4 Pt B):639-50. Antivir Ther. 2007. PMID: 17944271 Review.
Cited by
-
The role of viral population diversity in adaptation of bovine coronavirus to new host environments.PLoS One. 2013;8(1):e52752. doi: 10.1371/journal.pone.0052752. Epub 2013 Jan 7. PLoS One. 2013. PMID: 23308119 Free PMC article.
-
Heterogeneous expression of the SARS-Coronavirus-2 receptor ACE2 in the human respiratory tract.bioRxiv [Preprint]. 2020 Aug 13:2020.04.22.056127. doi: 10.1101/2020.04.22.056127. bioRxiv. 2020. Update in: EBioMedicine. 2020 Oct;60:102976. doi: 10.1016/j.ebiom.2020.102976 PMID: 32577664 Free PMC article. Updated. Preprint.
-
Mutations in membrane-fusion subunit of spike glycoprotein play crucial role in the recent outbreak of COVID-19.J Med Virol. 2021 May;93(5):2790-2798. doi: 10.1002/jmv.26598. Epub 2021 Feb 23. J Med Virol. 2021. PMID: 33090493 Free PMC article.
-
Integrative structural studies of the SARS-CoV-2 spike protein during the fusion process (2022).Curr Res Struct Biol. 2022;4:220-230. doi: 10.1016/j.crstbi.2022.06.004. Epub 2022 Jun 23. Curr Res Struct Biol. 2022. PMID: 35765663 Free PMC article.
-
Matrix metalloproteinases and tissue inhibitors of metalloproteinases in murine β-coronavirus-induced neuroinflammation.Virology. 2022 Jan;566:122-135. doi: 10.1016/j.virol.2021.11.012. Epub 2021 Dec 2. Virology. 2022. PMID: 34906793 Free PMC article.
References
-
- Beaufort N., et al. 2007. The human airway trypsin-like protease modulates the urokinase receptor (uPAR, CD87) structure and functions. Am. J. Physiol. Lung Cell. Mol. Physiol. 292:L1263–L1272 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

