Chlamydia trachomatis co-opts GBF1 and CERT to acquire host sphingomyelin for distinct roles during intracellular development
- PMID: 21909260
- PMCID: PMC3164637
- DOI: 10.1371/journal.ppat.1002198
Chlamydia trachomatis co-opts GBF1 and CERT to acquire host sphingomyelin for distinct roles during intracellular development
Erratum in
- PLoS Pathog. 2013 Aug;9(8). doi: 10.1371/annotation/f8e7c7e3-c347-4243-9146-db77900cb90c
Abstract
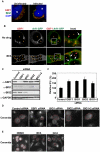
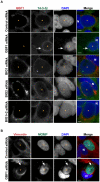
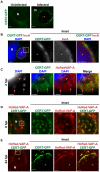
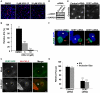
The strain designated Chlamydia trachomatis serovar that was used for experiments in this paper is Chlamydia muridarum, a species closely related to C. trachomatis (and formerly termed the Mouse Pneumonitis strain of C. trachomatis. [corrected]. The obligate intracellular pathogen Chlamydia trachomatis replicates within a membrane-bound inclusion that acquires host sphingomyelin (SM), a process that is essential for replication as well as inclusion biogenesis. Previous studies demonstrate that SM is acquired by a Brefeldin A (BFA)-sensitive vesicular trafficking pathway, although paradoxically, this pathway is dispensable for bacterial replication. This finding suggests that other lipid transport mechanisms are involved in the acquisition of host SM. In this work, we interrogated the role of specific components of BFA-sensitive and BFA-insensitive lipid trafficking pathways to define their contribution in SM acquisition during infection. We found that C. trachomatis hijacks components of both vesicular and non-vesicular lipid trafficking pathways for SM acquisition but that the SM obtained from these separate pathways is being utilized by the pathogen in different ways. We show that C. trachomatis selectively co-opts only one of the three known BFA targets, GBF1, a regulator of Arf1-dependent vesicular trafficking within the early secretory pathway for vesicle-mediated SM acquisition. The Arf1/GBF1-dependent pathway of SM acquisition is essential for inclusion membrane growth and stability but is not required for bacterial replication. In contrast, we show that C. trachomatis co-opts CERT, a lipid transfer protein that is a key component in non-vesicular ER to trans-Golgi trafficking of ceramide (the precursor for SM), for C. trachomatis replication. We demonstrate that C. trachomatis recruits CERT, its ER binding partner, VAP-A, and SM synthases, SMS1 and SMS2, to the inclusion and propose that these proteins establish an on-site SM biosynthetic factory at or near the inclusion. We hypothesize that SM acquired by CERT-dependent transport of ceramide and subsequent conversion to SM is necessary for C. trachomatis replication whereas SM acquired by the GBF1-dependent pathway is essential for inclusion growth and stability. Our results reveal a novel mechanism by which an intracellular pathogen redirects SM biosynthesis to its replicative niche.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures
Comment in
-
Rerouting of host lipids by bacteria: are you CERTain you need a vesicle?PLoS Pathog. 2011 Sep;7(9):e1002208. doi: 10.1371/journal.ppat.1002208. Epub 2011 Sep 1. PLoS Pathog. 2011. PMID: 21909265 Free PMC article. No abstract available.
Similar articles
-
Chlamydia trachomatis-infected human cells convert ceramide to sphingomyelin without sphingomyelin synthases 1 and 2.FEBS Lett. 2020 Feb;594(3):519-529. doi: 10.1002/1873-3468.13632. Epub 2019 Oct 22. FEBS Lett. 2020. PMID: 31596951
-
The lipid transfer protein CERT interacts with the Chlamydia inclusion protein IncD and participates to ER-Chlamydia inclusion membrane contact sites.PLoS Pathog. 2011 Jun;7(6):e1002092. doi: 10.1371/journal.ppat.1002092. Epub 2011 Jun 23. PLoS Pathog. 2011. PMID: 21731489 Free PMC article.
-
Rerouting of host lipids by bacteria: are you CERTain you need a vesicle?PLoS Pathog. 2011 Sep;7(9):e1002208. doi: 10.1371/journal.ppat.1002208. Epub 2011 Sep 1. PLoS Pathog. 2011. PMID: 21909265 Free PMC article. No abstract available.
-
Sphingolipid Metabolism and Transport in Chlamydia trachomatis and Chlamydia psittaci Infections.Front Cell Dev Biol. 2019 Oct 4;7:223. doi: 10.3389/fcell.2019.00223. eCollection 2019. Front Cell Dev Biol. 2019. PMID: 31637241 Free PMC article. Review.
-
CERT and intracellular trafficking of ceramide.Biochim Biophys Acta. 2007 Jun;1771(6):644-53. doi: 10.1016/j.bbalip.2007.01.009. Epub 2007 Jan 23. Biochim Biophys Acta. 2007. PMID: 17314061 Review.
Cited by
-
Isolation of Chlamydia trachomatis and membrane vesicles derived from host and bacteria.J Microbiol Methods. 2012 Nov;91(2):222-30. doi: 10.1016/j.mimet.2012.08.012. Epub 2012 Aug 30. J Microbiol Methods. 2012. PMID: 22960504 Free PMC article.
-
Trifunctional sphingomyelin derivatives enable nanoscale resolution of sphingomyelin turnover in physiological and infection processes via expansion microscopy.Nat Commun. 2024 Aug 28;15(1):7456. doi: 10.1038/s41467-024-51874-w. Nat Commun. 2024. PMID: 39198435 Free PMC article.
-
Proximity Labeling To Map Host-Pathogen Interactions at the Membrane of a Bacterium-Containing Vacuole in Chlamydia trachomatis-Infected Human Cells.Infect Immun. 2019 Oct 18;87(11):e00537-19. doi: 10.1128/IAI.00537-19. Print 2019 Nov. Infect Immun. 2019. PMID: 31405957 Free PMC article.
-
Targeting host lipid flows: Exploring new antiviral and antibiotic strategies.Cell Microbiol. 2019 Mar;21(3):e12996. doi: 10.1111/cmi.12996. Epub 2019 Jan 17. Cell Microbiol. 2019. PMID: 30585688 Free PMC article. Review.
-
Establishing the intracellular niche of obligate intracellular vacuolar pathogens.Front Cell Infect Microbiol. 2023 Aug 14;13:1206037. doi: 10.3389/fcimb.2023.1206037. eCollection 2023. Front Cell Infect Microbiol. 2023. PMID: 37645379 Free PMC article. Review.
References
-
- Schachter J. The intracellular life of Chlamydia. Curr Topics Microbiol Immunol. 1988;138:109–139. - PubMed
-
- Campbell LA, Kuo CC. Chlamydia pneumoniae and atherosclerosis. Semin Respir Infect. 2003;18:48–54. - PubMed
-
- Hackstadt T. Cell Biology. In: Stephens R, editor. Chlamydia. Washington, D.C.: ASM press; 1999. pp. 101–138.
Publication types
MeSH terms
Substances
Grants and funding
- CAPMC/ CIHR/Canada
- R01 GM079139/GM/NIGMS NIH HHS/United States
- R01-AI073770/AI/NIAID NIH HHS/United States
- T32 AI060537/AI/NIAID NIH HHS/United States
- T32AI007334/AI/NIAID NIH HHS/United States
- S10 RR026758/RR/NCRR NIH HHS/United States
- T32 AI007334/AI/NIAID NIH HHS/United States
- C06 RR16490/RR/NCRR NIH HHS/United States
- C06 RR016490/RR/NCRR NIH HHS/United States
- R01 AI073770/AI/NIAID NIH HHS/United States
- R01-GM079139/GM/NIGMS NIH HHS/United States
- T32AI060537/AI/NIAID NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

