Coordination of KSHV latent and lytic gene control by CTCF-cohesin mediated chromosome conformation
- PMID: 21876668
- PMCID: PMC3158054
- DOI: 10.1371/journal.ppat.1002140
Coordination of KSHV latent and lytic gene control by CTCF-cohesin mediated chromosome conformation
Abstract
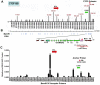
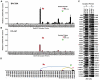
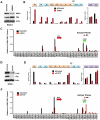
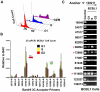
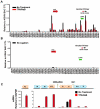
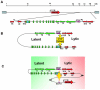
Herpesvirus persistence requires a dynamic balance between latent and lytic cycle gene expression, but how this balance is maintained remains enigmatic. We have previously shown that the Kaposi's Sarcoma-Associated Herpesvirus (KSHV) major latency transcripts encoding LANA, vCyclin, vFLIP, v-miRNAs, and Kaposin are regulated, in part, by a chromatin organizing element that binds CTCF and cohesins. Using viral genome-wide chromatin conformation capture (3C) methods, we now show that KSHV latency control region is physically linked to the promoter regulatory region for ORF50, which encodes the KSHV immediate early protein RTA. Other linkages were also observed, including an interaction between the 5' and 3' end of the latency transcription cluster. Mutation of the CTCF-cohesin binding site reduced or eliminated the chromatin conformation linkages, and deregulated viral transcription and genome copy number control. siRNA depletion of CTCF or cohesin subunits also disrupted chromosomal linkages and deregulated viral latent and lytic gene transcription. Furthermore, the linkage between the latent and lytic control region was subject to cell cycle fluctuation and disrupted during lytic cycle reactivation, suggesting that these interactions are dynamic and regulatory. Our findings indicate that KSHV genomes are organized into chromatin loops mediated by CTCF and cohesin interactions, and that these inter-chromosomal linkages coordinate latent and lytic gene control.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures


Similar articles
-
Complex Interactions between Cohesin and CTCF in Regulation of Kaposi's Sarcoma-Associated Herpesvirus Lytic Transcription.J Virol. 2020 Jan 6;94(2):e01279-19. doi: 10.1128/JVI.01279-19. Print 2020 Jan 6. J Virol. 2020. PMID: 31666380 Free PMC article.
-
Cohesins repress Kaposi's sarcoma-associated herpesvirus immediate early gene transcription during latency.J Virol. 2012 Sep;86(17):9454-64. doi: 10.1128/JVI.00787-12. Epub 2012 Jun 27. J Virol. 2012. PMID: 22740398 Free PMC article.
-
Cell cycle control of Kaposi's sarcoma-associated herpesvirus latency transcription by CTCF-cohesin interactions.J Virol. 2009 Jun;83(12):6199-210. doi: 10.1128/JVI.00052-09. Epub 2009 Apr 15. J Virol. 2009. PMID: 19369356 Free PMC article.
-
Lytic cycle switches of oncogenic human gammaherpesviruses.Adv Cancer Res. 2007;97:81-109. doi: 10.1016/S0065-230X(06)97004-3. Adv Cancer Res. 2007. PMID: 17419942 Review.
-
Molecular biology of KSHV lytic reactivation.Viruses. 2015 Jan 14;7(1):116-53. doi: 10.3390/v7010116. Viruses. 2015. PMID: 25594835 Free PMC article. Review.
Cited by
-
CTCF regulates Kaposi's sarcoma-associated herpesvirus latency transcription by nucleosome displacement and RNA polymerase programming.J Virol. 2013 Feb;87(3):1789-99. doi: 10.1128/JVI.02283-12. Epub 2012 Nov 28. J Virol. 2013. PMID: 23192870 Free PMC article.
-
Utilization of Host Cell Chromosome Conformation by Viral Pathogens: Knowing When to Hold and When to Fold.Front Immunol. 2021 Mar 25;12:633762. doi: 10.3389/fimmu.2021.633762. eCollection 2021. Front Immunol. 2021. PMID: 33841414 Free PMC article. Review.
-
Snapshots: chromatin control of viral infection.Virology. 2013 Jan 5;435(1):141-56. doi: 10.1016/j.virol.2012.09.023. Virology. 2013. PMID: 23217624 Free PMC article. Review.
-
Interpreting the Epstein-Barr Virus (EBV) epigenome using high-throughput data.Viruses. 2013 Apr 2;5(4):1042-54. doi: 10.3390/v5041042. Viruses. 2013. PMID: 23549386 Free PMC article. Review.
-
Complex Interactions between Cohesin and CTCF in Regulation of Kaposi's Sarcoma-Associated Herpesvirus Lytic Transcription.J Virol. 2020 Jan 6;94(2):e01279-19. doi: 10.1128/JVI.01279-19. Print 2020 Jan 6. J Virol. 2020. PMID: 31666380 Free PMC article.
References
-
- Chang Y, Cesarman E, Pessin MS, Lee F, Culpepper J, et al. Identification of herpesvirus-like DNA sequences in AIDS-associated Kaposi's sarcoma.[comment]. Science. 1994;266:1865–1869. - PubMed
-
- Cesarman E, Chang Y, Moore PS, Said JW, Knowles DM. Kaposi's sarcoma-associated herpesvirus-like DNA sequences in AIDS- related body-cavity-based lymphomas [see comments]. N Engl J Med. 1995;332:1186–1191. - PubMed
-
- Soulier J, Grollet L, Oksenhendler E, Cacoub P, Cazals-Hatem D, et al. Kaposi's sarcoma-associated herpesvirus-like DNA sequences in multicentric Castleman's disease. Blood. 1995;86:1276–1280. - PubMed
-
- Ganem D. KSHV infection and the pathogenesis of Kaposi's sarcoma. Annu Rev Pathol. 2006;1:273–296. - PubMed
-
- Schulz TF. The pleiotropic effects of Kaposi's sarcoma herpesvirus. J Pathol. 2006;208:187–198. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

