Noncoding RNA gene silencing through genomic integration of RNA destabilizing elements using zinc finger nucleases
- PMID: 21844124
- PMCID: PMC3205578
- DOI: 10.1101/gr.122358.111
Noncoding RNA gene silencing through genomic integration of RNA destabilizing elements using zinc finger nucleases
Abstract
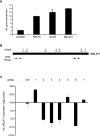
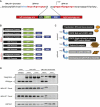
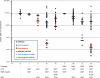
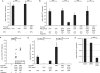
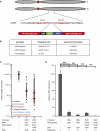
Zinc finger nucleases (ZFNs) allow site-specific manipulation of the genome. So far, the use of ZFNs to create gene knockouts has been restricted to protein-coding genes. However, non-protein-encoding RNAs (ncRNA) play important roles in the cell, although the functions of most ncRNAs are unknown. Here, we describe a ZFN-based method suited for the silencing of protein-coding and noncoding genes. This method relies on the ZFN-mediated integration of RNA destabilizing elements into the human genome, e.g., poly(A) signals functioning as termination elements and destabilizing downstream sequences. The biallelic integration of poly(A) signals into the gene locus of the long ncRNA MALAT1 resulted in a 1000-fold decrease of RNA expression. Thus, this approach is more specific and 300 times more efficient than RNA interference techniques. The opportunity to create a variety of loss-of-function tumor model cell lines in different cancer backgrounds will promote future functional analyses of important long noncoding RNA transcripts.
Figures

Similar articles
-
Silencing long noncoding RNAs with genome-editing tools.Methods Mol Biol. 2015;1239:241-50. doi: 10.1007/978-1-4939-1862-1_13. Methods Mol Biol. 2015. PMID: 25408410
-
Genome editing with CompoZr custom zinc finger nucleases (ZFNs).J Vis Exp. 2012 Jun 14;(64):e3304. doi: 10.3791/3304. J Vis Exp. 2012. PMID: 22732945 Free PMC article.
-
Rapid mutation of endogenous zebrafish genes using zinc finger nucleases made by Oligomerized Pool ENgineering (OPEN).PLoS One. 2009;4(2):e4348. doi: 10.1371/journal.pone.0004348. Epub 2009 Feb 9. PLoS One. 2009. PMID: 19198653 Free PMC article.
-
Advances in genetic modification of farm animals using zinc-finger nucleases (ZFN).Chromosome Res. 2015 Feb;23(1):7-15. doi: 10.1007/s10577-014-9451-7. Chromosome Res. 2015. PMID: 25596823 Review.
-
Cell Biology Symposium: Zinc finger nucleases to create custom-designed modifications in the swine (Sus scrofa) genome.J Anim Sci. 2012 Apr;90(4):1111-7. doi: 10.2527/jas.2011-4546. Epub 2011 Oct 28. J Anim Sci. 2012. PMID: 22038991 Review.
Cited by
-
LncRNA-mediated cartilage homeostasis in osteoarthritis: a narrative review.Front Med (Lausanne). 2024 Feb 21;11:1326843. doi: 10.3389/fmed.2024.1326843. eCollection 2024. Front Med (Lausanne). 2024. PMID: 38449881 Free PMC article. Review.
-
Targeting and engineering long non-coding RNAs for cancer therapy.Nat Rev Genet. 2024 Aug;25(8):578-595. doi: 10.1038/s41576-024-00693-2. Epub 2024 Feb 29. Nat Rev Genet. 2024. PMID: 38424237 Review.
-
Techniques for investigating lncRNA transcript functions in neurodevelopment.Mol Psychiatry. 2024 Apr;29(4):874-890. doi: 10.1038/s41380-023-02377-5. Epub 2023 Dec 25. Mol Psychiatry. 2024. PMID: 38145986 Free PMC article. Review.
-
Long Non-coding RNAs and CRISPR-Cas Edition in Tumorigenesis.Adv Exp Med Biol. 2023;1429:41-58. doi: 10.1007/978-3-031-33325-5_3. Adv Exp Med Biol. 2023. PMID: 37486515
-
Long non-coding RNAs modulate tumor microenvironment to promote metastasis: novel avenue for therapeutic intervention.Front Cell Dev Biol. 2023 Jun 13;11:1164301. doi: 10.3389/fcell.2023.1164301. eCollection 2023. Front Cell Dev Biol. 2023. PMID: 37384249 Free PMC article. Review.
References
-
- Carninci P, Kasukawa T, Katayama S, Gough J, Frith MC, Maeda N, Oyama R, Ravasi T, Lenhard B, Wells C, et al. 2005. The transcriptional landscape of the mammalian genome. Science 309: 1559–1563 - PubMed
-
- Elbashir SM, Harborth J, Lendeckel W, Yalcin A, Weber K, Tuschl T 2001. Duplexes of 21-nucleotide RNAs mediate RNA interference in cultured mammalian cells. Nature 411: 494–498 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
