Protein-protein fusion catalyzed by sortase A
- PMID: 21494692
- PMCID: PMC3071835
- DOI: 10.1371/journal.pone.0018342
Protein-protein fusion catalyzed by sortase A
Abstract
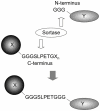
Chimeric proteins boast widespread use in areas ranging from cell biology to drug delivery. Post-translational protein fusion using the bacterial transpeptidase sortase A provides an attractive alternative when traditional gene fusion fails. We describe use of this enzyme for in vitro protein ligation and report the successful fusion of 10 pairs of protein domains with preserved functionality--demonstrating the robust and facile nature of this reaction.
Conflict of interest statement
Figures



Similar articles
-
Making and breaking peptide bonds: protein engineering using sortase.Angew Chem Int Ed Engl. 2011 May 23;50(22):5024-32. doi: 10.1002/anie.201008267. Epub 2011 Apr 27. Angew Chem Int Ed Engl. 2011. PMID: 21538739 Review.
-
Enhancement of sortase A-mediated protein ligation by inducing a β-hairpin structure around the ligation site.Chem Commun (Camb). 2011 Apr 28;47(16):4742-4. doi: 10.1039/c0cc05334a. Epub 2011 Mar 15. Chem Commun (Camb). 2011. PMID: 21409251
-
Observing selected domains in multi-domain proteins via sortase-mediated ligation and NMR spectroscopy.J Biomol NMR. 2011 Jan;49(1):3-7. doi: 10.1007/s10858-010-9464-2. Epub 2010 Dec 29. J Biomol NMR. 2011. PMID: 21188472 Free PMC article.
-
Group B Streptococcus pilus sortase regulation: a single mutation in the lid region induces pilin protein polymerization in vitro.FASEB J. 2013 Aug;27(8):3144-54. doi: 10.1096/fj.13-227793. Epub 2013 Apr 30. FASEB J. 2013. PMID: 23631841
-
Sortase-mediated ligation: a gift from Gram-positive bacteria to protein engineering.Chembiochem. 2009 Mar 23;10(5):787-98. doi: 10.1002/cbic.200800724. Chembiochem. 2009. PMID: 19199328 Review. No abstract available.
Cited by
-
Integrating single-molecule spectroscopy and simulations for the study of intrinsically disordered proteins.Methods. 2021 Sep;193:116-135. doi: 10.1016/j.ymeth.2021.03.018. Epub 2021 Apr 6. Methods. 2021. PMID: 33831596 Free PMC article.
-
Greatest Hits-Innovative Technologies for High Throughput Identification of Bispecific Antibodies.Int J Mol Sci. 2020 Sep 8;21(18):6551. doi: 10.3390/ijms21186551. Int J Mol Sci. 2020. PMID: 32911608 Free PMC article. Review.
-
Antibody drug conjugates: design and selection of linker, payload and conjugation chemistry.AAPS J. 2015 Mar;17(2):339-51. doi: 10.1208/s12248-014-9710-8. Epub 2015 Jan 22. AAPS J. 2015. PMID: 25604608 Free PMC article. Review.
-
Improving the secretion of a methyl parathion hydrolase in Pichia pastoris by modifying its N-terminal sequence.PLoS One. 2014 May 7;9(5):e96974. doi: 10.1371/journal.pone.0096974. eCollection 2014. PLoS One. 2014. PMID: 24806460 Free PMC article.
-
Semienzymatic cyclization of disulfide-rich peptides using Sortase A.J Biol Chem. 2014 Mar 7;289(10):6627-6638. doi: 10.1074/jbc.M113.539262. Epub 2014 Jan 14. J Biol Chem. 2014. PMID: 24425873 Free PMC article.
References
-
- Dawson PE, Kent SB. Synthesis of native proteins by chemical ligation. Annu Rev Biochem. 2000;69:923–960. - PubMed
-
- Blaschke UK, Silberstein J, Muir TW. Protein engineering by expressed protein ligation. Methods Enzymol. 2000;328:478–496. - PubMed
-
- Jackson DY, Burnier J, Quan C, Stanley M, Tom J, et al. A designed peptide ligase for total synthesis of ribonuclease A with unnatural catalytic residues. Science. 1994;266:243–247. - PubMed
-
- Ayers B, Blaschke UK, Camarero JA, Cotton GJ, Holford M, et al. Introduction of unnatural amino acids into proteins using expressed protein ligation. Biopolymers. 1999;51:343–354. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

