Infection of primary human tonsillar lymphoid cells by KSHV reveals frequent but abortive infection of T cells
- PMID: 21353276
- PMCID: PMC3070441
- DOI: 10.1016/j.virol.2010.12.036
Infection of primary human tonsillar lymphoid cells by KSHV reveals frequent but abortive infection of T cells
Abstract
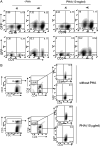
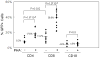
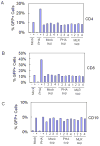
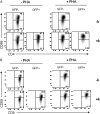
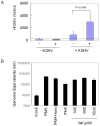
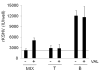
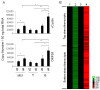
The lymphotropic herpesvirus KSHV principally infects B cells in vivo and is linked to several human B cell lymphoproliferative syndromes. Here we examine the susceptibility of primary tonsillar lymphocytes to infection by a recombinant KSHV (rKSHV.219) that constitutively expresses GFP. At an MOI of ~1, ca. 5-10% of CD19+ B cells became GFP-positive. Surprisingly, in the same culture many more T cells became infected. However, in contrast to isolated B cells, isolated infected T cells did not support correct viral transcription and did not produce infectious virus, indicating the presence of one or more post-entry blocks to lytic KSHV replication in T cells. No immortalization or transformation has yet been observed in either B or T cells. These results affirm the feasibility of studying KSHV infection in primary lymphoid cells, and help to rationalize the detection of KSHV DNA in rare human T cell lymphomas in vivo.
Copyright © 2010 Elsevier Inc. All rights reserved.
Figures


Similar articles
-
Transcriptome analysis of Kaposi's sarcoma-associated herpesvirus during de novo primary infection of human B and endothelial cells.J Virol. 2015 Mar;89(6):3093-111. doi: 10.1128/JVI.02507-14. Epub 2014 Dec 31. J Virol. 2015. PMID: 25552714 Free PMC article.
-
KSHV/HHV-8 infection of human hematopoietic progenitor (CD34+) cells: persistence of infection during hematopoiesis in vitro and in vivo.Blood. 2006 Jul 1;108(1):141-51. doi: 10.1182/blood-2005-04-1697. Epub 2006 Mar 16. Blood. 2006. PMID: 16543476 Free PMC article.
-
Use of the red fluorescent protein as a marker of Kaposi's sarcoma-associated herpesvirus lytic gene expression.Virology. 2004 Aug 1;325(2):225-40. doi: 10.1016/j.virol.2004.03.049. Virology. 2004. PMID: 15246263
-
Primary lymphocyte infection models for KSHV and its putative tumorigenesis mechanisms in B cell lymphomas.J Microbiol. 2017 May;55(5):319-329. doi: 10.1007/s12275-017-7075-2. Epub 2017 Apr 29. J Microbiol. 2017. PMID: 28455586 Review.
-
EBV and KSHV Infection Dysregulates Autophagy to Optimize Viral Replication, Prevent Immune Recognition and Promote Tumorigenesis.Viruses. 2018 Oct 31;10(11):599. doi: 10.3390/v10110599. Viruses. 2018. PMID: 30384495 Free PMC article. Review.
Cited by
-
Towards Better Understanding of KSHV Life Cycle: from Transcription and Posttranscriptional Regulations to Pathogenesis.Virol Sin. 2019 Apr;34(2):135-161. doi: 10.1007/s12250-019-00114-3. Epub 2019 Apr 25. Virol Sin. 2019. PMID: 31025296 Free PMC article. Review.
-
Kaposi's sarcoma-associated herpesvirus-encoded microRNA miR-K12-11 attenuates transforming growth factor beta signaling through suppression of SMAD5.J Virol. 2012 Feb;86(3):1372-81. doi: 10.1128/JVI.06245-11. Epub 2011 Oct 19. J Virol. 2012. PMID: 22013049 Free PMC article.
-
Infection of lymphoblastoid cell lines by Kaposi's sarcoma-associated herpesvirus: critical role of cell-associated virus.J Virol. 2011 Oct;85(19):9767-77. doi: 10.1128/JVI.05136-11. Epub 2011 Jul 27. J Virol. 2011. PMID: 21795352 Free PMC article.
-
KSHV induces immunoglobulin rearrangements in mature B lymphocytes.PLoS Pathog. 2018 Apr 16;14(4):e1006967. doi: 10.1371/journal.ppat.1006967. eCollection 2018 Apr. PLoS Pathog. 2018. PMID: 29659614 Free PMC article.
-
Variable episomal silencing of a recombinant herpesvirus renders its encoded GFP an unreliable marker of infection in primary cells.PLoS One. 2014 Nov 17;9(11):e111502. doi: 10.1371/journal.pone.0111502. eCollection 2014. PLoS One. 2014. PMID: 25402328 Free PMC article.
References
-
- Aman P, Gordon J, Lewin N, Nordstrom M, Ehlin-Henriksson B, Klein G, Carstensson A. Surface marker characterization of EBV target cells in normal blood and tonsil B lymphocyte populations. J Immunol. 1985;135(4):2362–7. - PubMed
-
- Ambroziak JA, Blackbourn DJ, Herndier BG, Glogau RG, Gullett JH, McDonald AR, Lennette ET, Levy JA. Herpes-like sequences in HIV-infected and uninfected Kaposi’s sarcoma patients. Science. 1995;268(5210):582–3. - PubMed
-
- An J, Sun Y, Sun R, Rettig MB. Kaposi’s sarcoma-associated herpesvirus encoded vFLIP induces cellular IL-6 expression: the role of the NF-kappaB and JNK/AP1 pathways. Oncogene. 2003;22(22):3371–85. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources

