Virus-encoded microRNAs
- PMID: 21277611
- PMCID: PMC3052296
- DOI: 10.1016/j.virol.2011.01.002
Virus-encoded microRNAs
Abstract
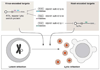
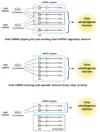
MicroRNAs (miRNAs) are the subject of enormous interest. They are small non-coding RNAs that play a regulatory role in numerous and diverse cellular processes such as immune function, apoptosis and tumorigenesis. Several virus families have been shown to encode miRNAs, and an appreciation for their roles in the viral infectious cycle continues to grow. Despite the identification of numerous (>225) viral miRNAs, an in depth functional understanding of most virus-encoded miRNAs is lacking. Here we focus on a few viral miRNAs with well-defined functions. We use these examples to extrapolate general themes of viral miRNA activities including autoregulation of viral gene expression, avoidance of host defenses, and a likely important role in maintaining latent and persistent infections. We hypothesize that although the molecular mechanisms and machinery are similar, the majority of viral miRNAs may utilize a target strategy that differs from host miRNAs. That is, many viral miRNAs may have evolved to regulate viral-encoded transcripts or networks of host genes that are unique to viral miRNAs. Included in this latter category is a likely abundant class of viral miRNAs that may regulate only one or a few principal host genes. Key steps forward for the field are discussed, including the need for additional functional studies that utilize surgical viral miRNA mutants combined with relevant models of infection.
Copyright © 2011 Elsevier Inc. All rights reserved.
Figures

Similar articles
-
Virus-encoded microRNAs: an overview and a look to the future.PLoS Pathog. 2012 Dec;8(12):e1003018. doi: 10.1371/journal.ppat.1003018. Epub 2012 Dec 20. PLoS Pathog. 2012. PMID: 23308061 Free PMC article. Review.
-
A human herpesvirus 6A-encoded microRNA: role in viral lytic replication.J Virol. 2015 Mar;89(5):2615-27. doi: 10.1128/JVI.02007-14. Epub 2014 Dec 17. J Virol. 2015. PMID: 25520507 Free PMC article.
-
Widespread evidence of viral miRNAs targeting host pathways.BMC Bioinformatics. 2013;14 Suppl 2(Suppl 2):S3. doi: 10.1186/1471-2105-14-S2-S3. Epub 2013 Jan 21. BMC Bioinformatics. 2013. PMID: 23369080 Free PMC article.
-
MicroRNAs and viral infection.Mol Cell. 2005 Oct 7;20(1):3-7. doi: 10.1016/j.molcel.2005.09.012. Mol Cell. 2005. PMID: 16209940 Review.
-
Novel microRNA-like viral small regulatory RNAs arising during human hepatitis A virus infection.FASEB J. 2014 Oct;28(10):4381-93. doi: 10.1096/fj.14-253534. Epub 2014 Jul 1. FASEB J. 2014. PMID: 25002121
Cited by
-
The small noncoding RNAs (sncRNAs) of murine gammaherpesvirus 68 (MHV-68) are involved in regulating the latent-to-lytic switch in vivo.Sci Rep. 2016 Aug 26;6:32128. doi: 10.1038/srep32128. Sci Rep. 2016. PMID: 27561205 Free PMC article.
-
The Role of microRNAs in the Pathogenesis of Herpesvirus Infection.Viruses. 2016 Jun 2;8(6):156. doi: 10.3390/v8060156. Viruses. 2016. PMID: 27271654 Free PMC article. Review.
-
Particulate Air Pollution Exposure and Expression of Viral and Human MicroRNAs in Blood: The Beijing Truck Driver Air Pollution Study.Environ Health Perspect. 2016 Mar;124(3):344-50. doi: 10.1289/ehp.1408519. Epub 2015 Jun 12. Environ Health Perspect. 2016. PMID: 26068961 Free PMC article.
-
Biology of Polyomavirus miRNA.Front Microbiol. 2021 Apr 6;12:662892. doi: 10.3389/fmicb.2021.662892. eCollection 2021. Front Microbiol. 2021. PMID: 33889147 Free PMC article. Review.
-
MicroRNA expression by an oncogenic retrovirus.Proc Natl Acad Sci U S A. 2012 Feb 21;109(8):2695-6. doi: 10.1073/pnas.1200328109. Epub 2012 Feb 2. Proc Natl Acad Sci U S A. 2012. PMID: 22308505 Free PMC article. No abstract available.
References
-
- Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–297. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

