Epstein-Barr Virus nuclear antigen 1 (EBNA1) confers resistance to apoptosis in EBV-positive B-lymphoma cells through up-regulation of survivin
- PMID: 21093004
- PMCID: PMC4287362
- DOI: 10.1016/j.virol.2010.10.029
Epstein-Barr Virus nuclear antigen 1 (EBNA1) confers resistance to apoptosis in EBV-positive B-lymphoma cells through up-regulation of survivin
Abstract
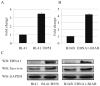
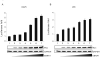
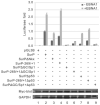
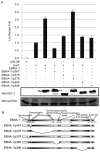
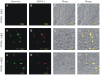
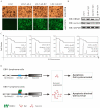
Resistance to apoptosis is an important component of the overall mechanism which drives the tumorigenic process. EBV is a ubiquitous human gamma-herpesvirus which preferentially establishes latent infection in viral infected B-lymphocytes. EBNA1 is typically expressed in most forms of EBV-positive malignancies and is important for replication of the latent episome in concert with replication of the host cells. Here, we investigate the effects of EBNA1 on survivin up-regulation in EBV-infected human B-lymphoma cells. We present evidence which demonstrates that EBNA1 forms a complex with Sp1 or Sp1-like proteins bound to their cis-element at the survivin promoter. This enhances the activity of the complex and up-regulates survivin. Knockdown of survivin and EBNA1 showed enhanced apoptosis in infected cells and thus supports a role for EBNA1 in suppressing apoptosis in EBV-infected cells. Here, we suggest that EBV encoded EBNA1 can contribute to the oncogenic process by up-regulating the apoptosis suppressor protein, survivin in EBV-associated B-lymphoma cells.
Copyright © 2010 Elsevier Inc. All rights reserved.
Figures


Similar articles
-
Carcinoma-risk variant of EBNA1 deregulates Epstein-Barr Virus episomal latency.Oncotarget. 2017 Jan 31;8(5):7248-7264. doi: 10.18632/oncotarget.14540. Oncotarget. 2017. PMID: 28077791 Free PMC article.
-
Baicalein inhibits growth of Epstein-Barr virus-positive nasopharyngeal carcinoma by repressing the activity of EBNA1 Q-promoter.Biomed Pharmacother. 2018 Jun;102:1003-1014. doi: 10.1016/j.biopha.2018.03.114. Epub 2018 Apr 5. Biomed Pharmacother. 2018. PMID: 29710517
-
Identification of MEF2B, EBF1, and IL6R as Direct Gene Targets of Epstein-Barr Virus (EBV) Nuclear Antigen 1 Critical for EBV-Infected B-Lymphocyte Survival.J Virol. 2015 Oct 14;90(1):345-55. doi: 10.1128/JVI.02318-15. Print 2016 Jan 1. J Virol. 2015. PMID: 26468528 Free PMC article.
-
Epstein-Barr virus-encoded EBNA1 and ZEBRA: targets for therapeutic strategies against EBV-carrying cancers.J Pathol. 2015 Jan;235(2):334-41. doi: 10.1002/path.4431. J Pathol. 2015. PMID: 25186125 Review.
-
Contributions of Epstein-Barr nuclear antigen 1 (EBNA1) to cell immortalization and survival.Viruses. 2012 Sep;4(9):1537-1547. doi: 10.3390/v4091537. Epub 2012 Sep 13. Viruses. 2012. PMID: 23170171 Free PMC article. Review.
Cited by
-
Association between Epstein-Barr virus infection and chemoresistance to docetaxel in gastric carcinoma.Mol Cells. 2011 Aug;32(2):173-9. doi: 10.1007/s10059-011-0066-y. Epub 2011 May 27. Mol Cells. 2011. PMID: 21626300 Free PMC article.
-
The single RBP-Jkappa site within the LANA promoter is crucial for establishing Kaposi's sarcoma-associated herpesvirus latency during primary infection.J Virol. 2011 Jul;85(13):6148-61. doi: 10.1128/JVI.02608-10. Epub 2011 Apr 20. J Virol. 2011. PMID: 21507979 Free PMC article.
-
Human Oncoviruses and p53 Tumor Suppressor Pathway Deregulation at the Origin of Human Cancers.Cancers (Basel). 2018 Jun 22;10(7):213. doi: 10.3390/cancers10070213. Cancers (Basel). 2018. PMID: 29932446 Free PMC article. Review.
-
EBV and Apoptosis: The Viral Master Regulator of Cell Fate?Viruses. 2017 Nov 13;9(11):339. doi: 10.3390/v9110339. Viruses. 2017. PMID: 29137176 Free PMC article. Review.
-
Research landmarks on the 60th anniversary of Epstein-Barr virus.Sci China Life Sci. 2024 Nov 4. doi: 10.1007/s11427-024-2766-0. Online ahead of print. Sci China Life Sci. 2024. PMID: 39505801 Review.
References
-
- Altieri DC. Validating survivin as a cancer therapeutic target. Nat Rev Cancer. 2003;3(1):46–54. - PubMed
-
- Ambrosini G, Adida C, Altieri DC. A novel anti-apoptosis gene, survivin, expressed in cancer and lymphoma. Nat Med. 1997;3(8):917–21. - PubMed
-
- Boidot R, Vegran F, Jacob D, Chevrier S, Cadouot M, Feron O, Solary E, Lizard-Nacol S. The transcription factor GATA-1 is overexpressed in breast carcinomas and contributes to survivin upregulation via a promoter polymorphism. Oncogene. 2010;29(17):2577–84. - PubMed
-
- Borbely AA, Murvai M, Konya J, Beck Z, Gergely L, Li F, Veress G. Effects of human papillomavirus type 16 oncoproteins on survivin gene expression. J Gen Virol. 2006;87:287–94. Pt 2. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

