The STRING database in 2011: functional interaction networks of proteins, globally integrated and scored
- PMID: 21045058
- PMCID: PMC3013807
- DOI: 10.1093/nar/gkq973
The STRING database in 2011: functional interaction networks of proteins, globally integrated and scored
Abstract
An essential prerequisite for any systems-level understanding of cellular functions is to correctly uncover and annotate all functional interactions among proteins in the cell. Toward this goal, remarkable progress has been made in recent years, both in terms of experimental measurements and computational prediction techniques. However, public efforts to collect and present protein interaction information have struggled to keep up with the pace of interaction discovery, partly because protein-protein interaction information can be error-prone and require considerable effort to annotate. Here, we present an update on the online database resource Search Tool for the Retrieval of Interacting Genes (STRING); it provides uniquely comprehensive coverage and ease of access to both experimental as well as predicted interaction information. Interactions in STRING are provided with a confidence score, and accessory information such as protein domains and 3D structures is made available, all within a stable and consistent identifier space. New features in STRING include an interactive network viewer that can cluster networks on demand, updated on-screen previews of structural information including homology models, extensive data updates and strongly improved connectivity and integration with third-party resources. Version 9.0 of STRING covers more than 1100 completely sequenced organisms; the resource can be reached at http://string-db.org.
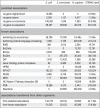
Figures



Similar articles
-
STRING v9.1: protein-protein interaction networks, with increased coverage and integration.Nucleic Acids Res. 2013 Jan;41(Database issue):D808-15. doi: 10.1093/nar/gks1094. Epub 2012 Nov 29. Nucleic Acids Res. 2013. PMID: 23203871 Free PMC article.
-
STRING 8--a global view on proteins and their functional interactions in 630 organisms.Nucleic Acids Res. 2009 Jan;37(Database issue):D412-6. doi: 10.1093/nar/gkn760. Epub 2008 Oct 21. Nucleic Acids Res. 2009. PMID: 18940858 Free PMC article.
-
STRING 7--recent developments in the integration and prediction of protein interactions.Nucleic Acids Res. 2007 Jan;35(Database issue):D358-62. doi: 10.1093/nar/gkl825. Epub 2006 Nov 10. Nucleic Acids Res. 2007. PMID: 17098935 Free PMC article.
-
Merging in-silico and in vitro salivary protein complex partners using the STRING database: A tutorial.J Proteomics. 2018 Jan 16;171:87-94. doi: 10.1016/j.jprot.2017.08.002. Epub 2017 Aug 3. J Proteomics. 2018. PMID: 28782718 Review.
-
GWIDD: a comprehensive resource for genome-wide structural modeling of protein-protein interactions.Hum Genomics. 2012 Jul 11;6(1):7. doi: 10.1186/1479-7364-6-7. Hum Genomics. 2012. PMID: 23245398 Free PMC article. Review.
Cited by
-
Identifying targets of the Sox domain protein Dichaete in the Drosophila CNS via targeted expression of dominant negative proteins.BMC Dev Biol. 2013 Jan 5;13:1. doi: 10.1186/1471-213X-13-1. BMC Dev Biol. 2013. PMID: 23289785 Free PMC article.
-
A multi-omics approach identifies key hubs associated with cell type-specific responses of airway epithelial cells to staphylococcal alpha-toxin.PLoS One. 2015 Mar 27;10(3):e0122089. doi: 10.1371/journal.pone.0122089. eCollection 2015. PLoS One. 2015. PMID: 25816343 Free PMC article.
-
Transcriptome profiling reveals response genes for downy mildew resistance in cucumber.Planta. 2021 Apr 29;253(5):112. doi: 10.1007/s00425-021-03603-6. Planta. 2021. PMID: 33914134
-
Functional diversity and structural disorder in the human ubiquitination pathway.PLoS One. 2013 May 29;8(5):e65443. doi: 10.1371/journal.pone.0065443. Print 2013. PLoS One. 2013. PMID: 23734257 Free PMC article.
-
The evolutionary dynamics of protein-protein interaction networks inferred from the reconstruction of ancient networks.PLoS One. 2013;8(3):e58134. doi: 10.1371/journal.pone.0058134. Epub 2013 Mar 20. PLoS One. 2013. PMID: 23526967 Free PMC article.
References
-
- Pujol A, Mosca R, Farres J, Aloy P. Unveiling the role of network and systems biology in drug discovery. Trends Pharmacol. Sci. 2010;31:115–123. - PubMed
-
- Klipp E, Wade RC, Kummer U. Biochemical network-based drug-target prediction. Curr. Opin. Biotechnol. 2010;21:511–516. - PubMed
-
- Janga SC, Diaz-Mejia JJ, Moreno-Hagelsieb G. Network-based function prediction and interactomics: The case for metabolic enzymes. Metab. Eng. 2010 - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

