Metabolic control of mitochondrial biogenesis through the PGC-1 family regulatory network
- PMID: 20933024
- PMCID: PMC3035754
- DOI: 10.1016/j.bbamcr.2010.09.019
Metabolic control of mitochondrial biogenesis through the PGC-1 family regulatory network
Abstract
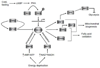
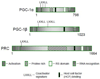
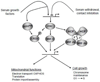
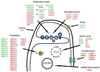
The PGC-1 family of regulated coactivators, consisting of PGC-1α, PGC-1β and PRC, plays a central role in a regulatory network governing the transcriptional control of mitochondrial biogenesis and respiratory function. These coactivators target multiple transcription factors including NRF-1, NRF-2 and the orphan nuclear hormone receptor, ERRα, among others. In addition, they themselves are the targets of coactivator and co-repressor complexes that regulate gene expression through chromatin remodeling. The expression of PGC-1 family members is modulated by extracellular signals controlling metabolism, differentiation or cell growth and in some cases their activities are known to be regulated by post-translational modification by the energy sensors, AMPK and SIRT1. Recent gene knockout and silencing studies of many members of the PGC-1 network have revealed phenotypes of wide ranging severity suggestive of complex compensatory interactions or broadly integrative functions that are not exclusive to mitochondrial biogenesis. The results point to a central role for the PGC-1 family in integrating mitochondrial biogenesis and energy production with many diverse cellular functions. This article is part of a Special Issue entitled: Mitochondria and Cardioprotection.
Copyright © 2010 Elsevier B.V. All rights reserved.
Figures

Similar articles
-
Control of mitochondrial transcription specificity factors (TFB1M and TFB2M) by nuclear respiratory factors (NRF-1 and NRF-2) and PGC-1 family coactivators.Mol Cell Biol. 2005 Feb;25(4):1354-66. doi: 10.1128/MCB.25.4.1354-1366.2005. Mol Cell Biol. 2005. PMID: 15684387 Free PMC article.
-
Estrogen-related receptor alpha directs peroxisome proliferator-activated receptor alpha signaling in the transcriptional control of energy metabolism in cardiac and skeletal muscle.Mol Cell Biol. 2004 Oct;24(20):9079-91. doi: 10.1128/MCB.24.20.9079-9091.2004. Mol Cell Biol. 2004. PMID: 15456881 Free PMC article.
-
The diabetes medication canagliflozin promotes mitochondrial remodelling of adipocyte via the AMPK-Sirt1-Pgc-1α signalling pathway.Adipocyte. 2020 Dec;9(1):484-494. doi: 10.1080/21623945.2020.1807850. Adipocyte. 2020. PMID: 32835596 Free PMC article.
-
Nuclear control of respiratory gene expression in mammalian cells.J Cell Biochem. 2006 Mar 1;97(4):673-83. doi: 10.1002/jcb.20743. J Cell Biochem. 2006. PMID: 16329141 Review.
-
PGC-1alpha, SIRT1 and AMPK, an energy sensing network that controls energy expenditure.Curr Opin Lipidol. 2009 Apr;20(2):98-105. doi: 10.1097/MOL.0b013e328328d0a4. Curr Opin Lipidol. 2009. PMID: 19276888 Free PMC article. Review.
Cited by
-
The clinical efficacy of cGMP-specific sildenafil on mitochondrial biogenesis induction and renal damage in cats with acute on chronic kidney disease.BMC Vet Res. 2024 Oct 31;20(1):499. doi: 10.1186/s12917-024-04345-9. BMC Vet Res. 2024. PMID: 39478527 Free PMC article.
-
Neohesperidin enhances PGC-1α-mediated mitochondrial biogenesis and alleviates hepatic steatosis in high fat diet fed mice.Nutr Diabetes. 2020 Aug 5;10(1):27. doi: 10.1038/s41387-020-00130-3. Nutr Diabetes. 2020. PMID: 32759940 Free PMC article.
-
Defective mitochondrial morphology and bioenergetic function in mice lacking the transcription factor Yin Yang 1 in skeletal muscle.Mol Cell Biol. 2012 Aug;32(16):3333-46. doi: 10.1128/MCB.00337-12. Epub 2012 Jun 18. Mol Cell Biol. 2012. PMID: 22711985 Free PMC article.
-
The Role of Epigenetic Control of Mitochondrial (Dys)Function in MASLD Onset and Progression.Nutrients. 2023 Nov 12;15(22):4757. doi: 10.3390/nu15224757. Nutrients. 2023. PMID: 38004151 Free PMC article. Review.
-
Molecular mechanisms of neonatal brain injury.Neurol Res Int. 2012;2012:506320. doi: 10.1155/2012/506320. Epub 2012 Jan 26. Neurol Res Int. 2012. PMID: 22363841 Free PMC article.
References
-
- Hatefi Y. The mitochondrial electron transport chain and oxidative phosphorylation system. Annu. Rev. Biochem. 1985;54:1015–1069. - PubMed
-
- Balaban RS. Regulation of oxidative phosphorylation in the mammalian cell. Am. J. Physiol. Cell Physiol. 1990;258:C377–C389. - PubMed
-
- Gray MW, Burgess G, Lang BF. Mitochondrial evolution. Science. 1999;283:1476–1481. - PubMed
-
- Taanman JW. The mitochondrial genome: structure, transcription, translation and replication. Biochim. Biophys. Acta Bio-Energetics. 1999;1410:103–123. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

