Genomic characterization of severe acute respiratory syndrome-related coronavirus in European bats and classification of coronaviruses based on partial RNA-dependent RNA polymerase gene sequences
- PMID: 20686038
- PMCID: PMC2953168
- DOI: 10.1128/JVI.00650-10
Genomic characterization of severe acute respiratory syndrome-related coronavirus in European bats and classification of coronaviruses based on partial RNA-dependent RNA polymerase gene sequences
Abstract
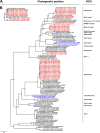
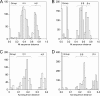
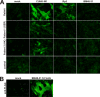
Bats may host emerging viruses, including coronaviruses (CoV). We conducted an evaluation of CoV in rhinolophid and vespertilionid bat species common in Europe. Rhinolophids carried severe acute respiratory syndrome (SARS)-related CoV at high frequencies and concentrations (26% of animals are positive; up to 2.4×10(8) copies per gram of feces), as well as two Alphacoronavirus clades, one novel and one related to the HKU2 clade. All three clades present in Miniopterus bats in China (HKU7, HKU8, and 1A related) were also present in European Miniopterus bats. An additional novel Alphacoronavirus clade (bat CoV [BtCoV]/BNM98-30) was detected in Nyctalus leisleri. A CoV grouping criterion was developed by comparing amino acid identities across an 816-bp fragment of the RNA-dependent RNA polymerases (RdRp) of all accepted mammalian CoV species (RdRp-based grouping units [RGU]). Criteria for defining separate RGU in mammalian CoV were a >4.8% amino acid distance for alphacoronaviruses and a >6.3% distance for betacoronaviruses. All the above-mentioned novel clades represented independent RGU. Strict associations between CoV RGU and host bat genera were confirmed for six independent RGU represented simultaneously in China and Europe. A SARS-related virus (BtCoV/BM48-31/Bulgaria/2008) from a Rhinolophus blasii (Rhi bla) bat was fully sequenced. It is predicted that proteins 3b and 6 were highly divergent from those proteins in all known SARS-related CoV. Open reading frame 8 (ORF8) was surprisingly absent. Surface expression of spike and staining with sera of SARS survivors suggested low antigenic overlap with SARS CoV. However, the receptor binding domain of SARS CoV showed higher similarity with that of BtCoV/BM48-31/Bulgaria/2008 than with that of any Chinese bat-borne CoV. Critical spike domains 472 and 487 were identical and similar, respectively. This study underlines the importance of assessments of the zoonotic potential of widely distributed bat-borne CoV.
Figures

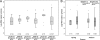

Similar articles
-
Coexistence of multiple coronaviruses in several bat colonies in an abandoned mineshaft.Virol Sin. 2016 Feb;31(1):31-40. doi: 10.1007/s12250-016-3713-9. Epub 2016 Feb 18. Virol Sin. 2016. PMID: 26920708 Free PMC article.
-
Severe Acute Respiratory Syndrome (SARS) Coronavirus ORF8 Protein Is Acquired from SARS-Related Coronavirus from Greater Horseshoe Bats through Recombination.J Virol. 2015 Oct;89(20):10532-47. doi: 10.1128/JVI.01048-15. Epub 2015 Aug 12. J Virol. 2015. PMID: 26269185 Free PMC article.
-
Novel Alphacoronaviruses and Paramyxoviruses Cocirculate with Type 1 and Severe Acute Respiratory System (SARS)-Related Betacoronaviruses in Synanthropic Bats of Luxembourg.Appl Environ Microbiol. 2017 Aug 31;83(18):e01326-17. doi: 10.1128/AEM.01326-17. Print 2017 Sep 15. Appl Environ Microbiol. 2017. PMID: 28710271 Free PMC article.
-
Ecology, evolution and classification of bat coronaviruses in the aftermath of SARS.Antiviral Res. 2014 Jan;101:45-56. doi: 10.1016/j.antiviral.2013.10.013. Epub 2013 Oct 31. Antiviral Res. 2014. PMID: 24184128 Free PMC article. Review.
-
Molecular epidemiology, evolution and phylogeny of SARS coronavirus.Infect Genet Evol. 2019 Jul;71:21-30. doi: 10.1016/j.meegid.2019.03.001. Epub 2019 Mar 4. Infect Genet Evol. 2019. PMID: 30844511 Free PMC article. Review.
Cited by
-
Evolutionary Arms Race between Virus and Host Drives Genetic Diversity in Bat Severe Acute Respiratory Syndrome-Related Coronavirus Spike Genes.J Virol. 2020 Sep 29;94(20):e00902-20. doi: 10.1128/JVI.00902-20. Print 2020 Sep 29. J Virol. 2020. PMID: 32699095 Free PMC article.
-
A Bat-Derived Putative Cross-Family Recombinant Coronavirus with a Reovirus Gene.PLoS Pathog. 2016 Sep 27;12(9):e1005883. doi: 10.1371/journal.ppat.1005883. eCollection 2016 Sep. PLoS Pathog. 2016. PMID: 27676249 Free PMC article.
-
Structural Genomic Analysis of SARS-CoV-2 and Other Coronaviruses.Front Genet. 2022 Apr 8;13:801902. doi: 10.3389/fgene.2022.801902. eCollection 2022. Front Genet. 2022. PMID: 35464844 Free PMC article.
-
A Cross Sectional Sampling Reveals Novel Coronaviruses in Bat Populations of Georgia.Viruses. 2021 Dec 31;14(1):72. doi: 10.3390/v14010072. Viruses. 2021. PMID: 35062276 Free PMC article.
-
SARS-like Coronaviruses in Horseshoe Bats (Rhinolophus spp.) in Russia, 2020.Viruses. 2022 Jan 9;14(1):113. doi: 10.3390/v14010113. Viruses. 2022. PMID: 35062318 Free PMC article.
References
-
- Becker, M. M., R. L. Graham, E. F. Donaldson, B. Rockx, A. C. Sims, T. Sheahan, R. J. Pickles, D. Corti, R. E. Johnston, R. S. Baric, and M. R. Denison. 2008. Synthetic recombinant bat SARS-like coronavirus is infectious in cultured cells and in mice. Proc. Natl. Acad. Sci. U. S. A. 105:19944-19949. - PMC - PubMed
-
- Benda, P., T. Ivanova, I. Horáček, V. Hanák, J. Červený, J. Gaisler, A. Gueorguieva, B. Petrov, and V. Vohralík. 2003. Bats (Mammalia: Chiroptera) of the eastern Mediterranean. Part 3. Review of bat distribution in Bulgaria. Acta Soc. Zool. Bohem. 67:245-357.
-
- Cavanagh, D. 1997. Nidovirales: a new order comprising Coronaviridae and Arteriviridae. Arch. Virol. 142:629-633. - PubMed
-
- Chinese Sars Molecular Epidemiology Consortium. 2004. Molecular evolution of the SARS coronavirus during the course of the SARS epidemic in China. Science 303:1666-1669. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

